R语言ggplot2科研数据数据可视化实用手册~第八章热图(heatmap)
R语言ggplot2科研数据数据可视化实用手册~第八章热图(heatmap)
用户7010445
发布于 2023-01-06 20:04:31
发布于 2023-01-06 20:04:31
Chapter 8 R语言ggplot2热图
今天下午7点到9点直播讲解如下代码,腾讯会议,感兴趣的参加,给推文打赏10元获取腾讯会议直播链接
8.1 pheatmap热图
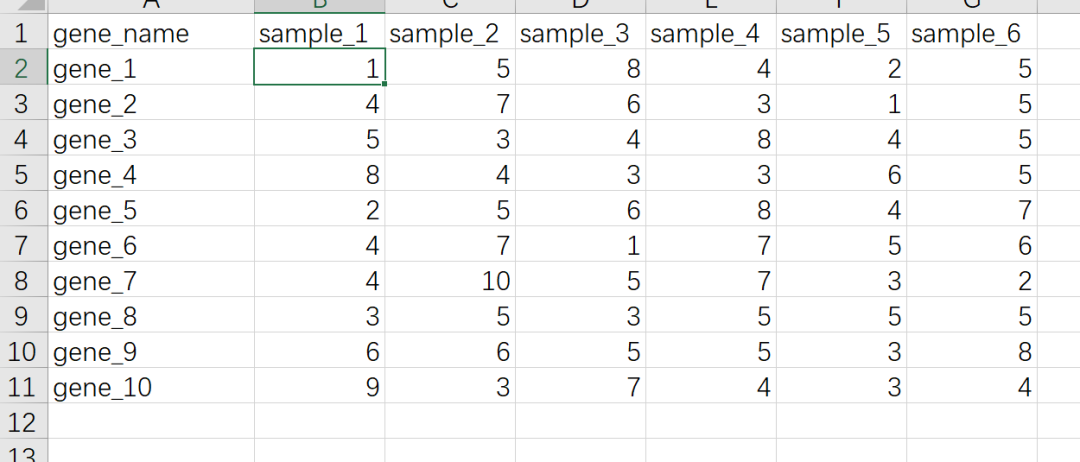
R语言里做热图最快捷的方式是用pheatmap这个R包,优点是用非常少的代码就可以出一个比较好看的图,缺点是细节修改不是很方便,比如要用热图展示基因表达量的数据,准备数据的格式如下
pheatmap不是R语言自带的R包,第一次使用需要先安装,安装直接使用命令install.packages("pheatmap")
读取数据
library(readxl)dat01<-read_excel("example_data/06-lineplot/dat08.xlsx")这里需要注意的 一个点是热图数据通常需要把第一列的基因名作为整个数据的行名,但是读取excel的函数好像没有指定列为行名的函数,当然可以将数据集读取进来以后再进行转换,另外一种方式就是把数据另存为csv格式,然后用读取csv格式数据的函数
这里需要注意的一点是转换为csv格式的时候选择如下截图中的2,不要选择1
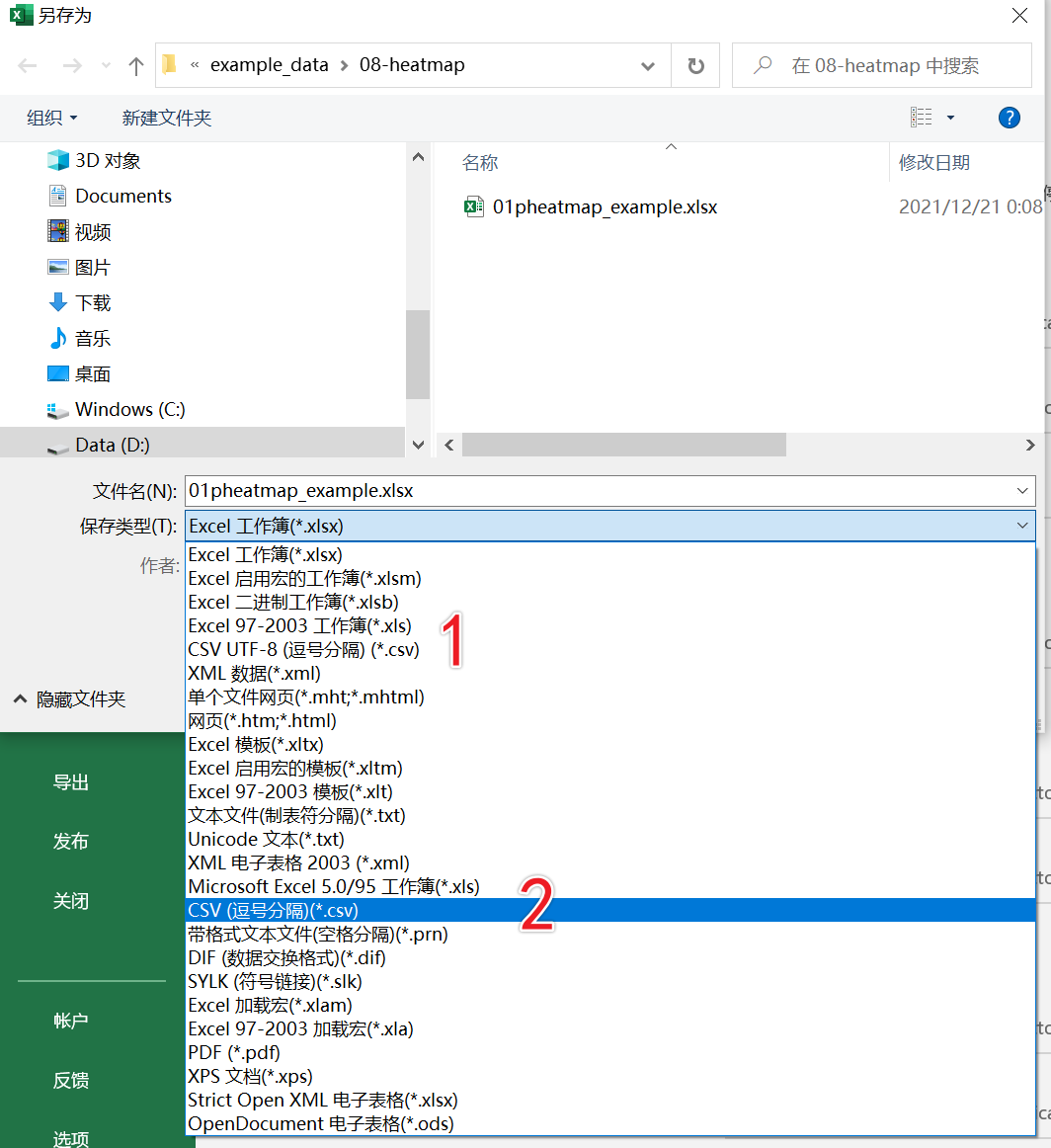
读取csv格式数据
dat01<-read.delim(file = "example_data/08-heatmap/01pheatmap_example.csv", header=TRUE, sep=",", row.names = 1)作图代码
dat01<-read.delim(file = "example_data/08-heatmap/01pheatmap_example.csv", header=TRUE, sep=",", row.names = 1)library(pheatmap)pheatmap(dat01)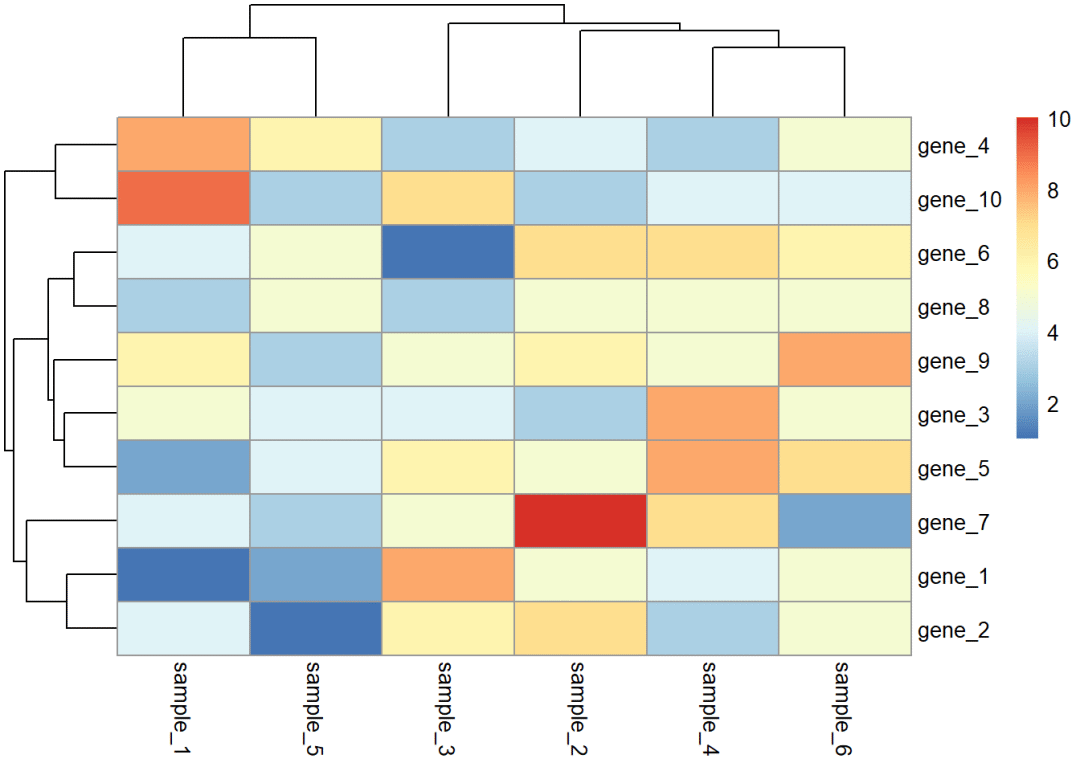
如果要在色块上添加文本,再单独准备一个和热图数据格式一样的数据,然后用display_numbers参数添加文本,这里我就直接使用热图的数据
dat01<-read.delim(file = "example_data/08-heatmap/01pheatmap_example.csv", header=TRUE, sep=",", row.names = 1)library(pheatmap)pheatmap(dat01)
pheatmap(dat01, display_numbers = dat01)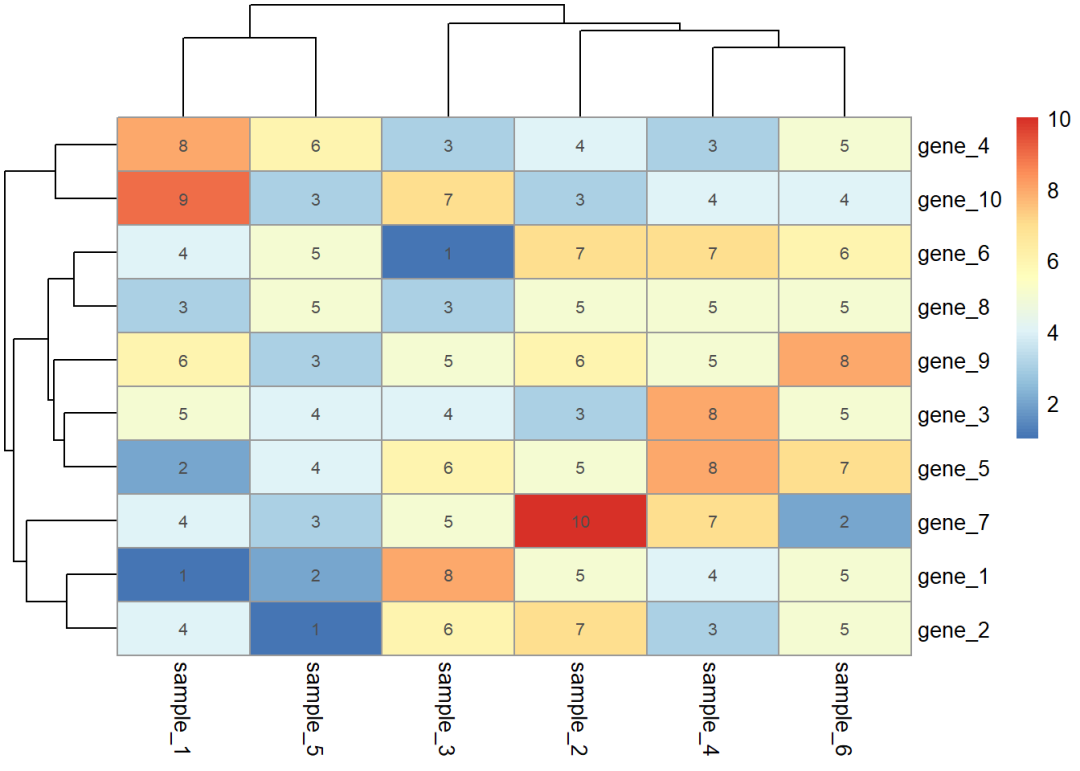
以上是对pheatmap这个R包的简要介绍
ggplot2也有直接做热图的函数 geom_tile(),ggplot2做热图可能代码稍微繁琐,但是优点是细节调整方便,基本上所有的细节都可以用代码来调整
ggplot2做热图还需要掌握的一个知识点是 长格式数据 和 宽格式 数据,ggplot2作图的输入数据都是长格式数据,长格式数据如下,一列x,一列y,还有一个数据
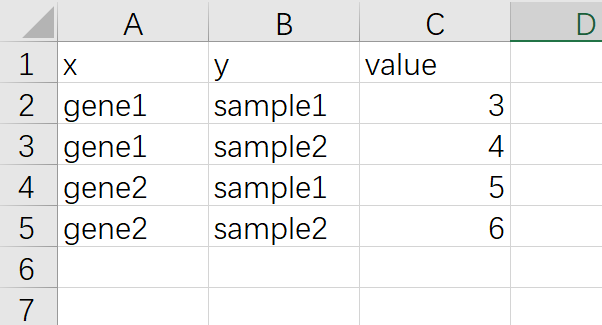
宽格式数据截图如下
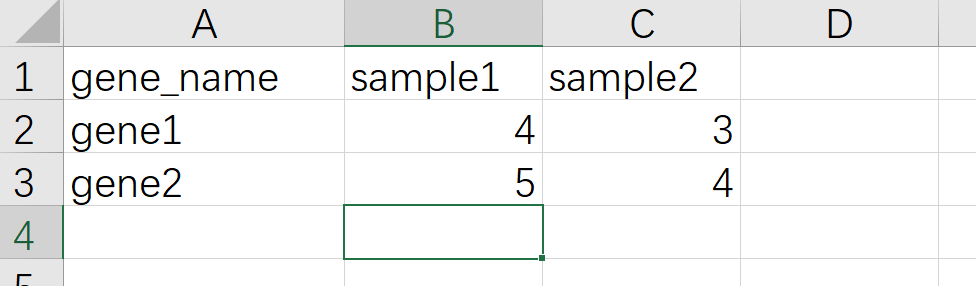
这个长宽格式转化是ggplot2作图必须理解的一个概念
R语言里提供了长宽格式数据互相转化的函数,这里我以tidyverse这个R包里的函数作为介绍,tidyverse主要是用来在数据处理的,也不是R语言自带的R包,需要运行安装命令install.packages("tidyverse")
宽格式数据转换为长格式用到的函数是pivot_longer()
转换代码
library(readxl)dat01<-read_excel("example_data/08-heatmap/02_wide_data.xlsx")head(dat01)## # A tibble: 2 x 3
## gene_name sample1 sample2
## <chr> <dbl> <dbl>
## 1 gene1 4 3
## 2 gene2 5 4library(tidyverse)dat01 %>% pivot_longer(-gene_name,names_to = "A",values_to = "B")## # A tibble: 4 x 3
## gene_name A B
## <chr> <chr> <dbl>
## 1 gene1 sample1 4
## 2 gene1 sample2 3
## 3 gene2 sample1 5
## 4 gene2 sample2 4长格式转换为宽格式的函数是pivot_wider()
转换代码
library(readxl)dat01<-read_excel("example_data/08-heatmap/02_long_data.xlsx")head(dat01)## # A tibble: 4 x 3
## x y value
## <chr> <chr> <dbl>
## 1 gene1 sample1 3
## 2 gene1 sample2 4
## 3 gene2 sample1 5
## 4 gene2 sample2 6library(tidyverse)dat01 %>% pivot_wider(names_from = y,values_from = value)## # A tibble: 2 x 3
## x sample1 sample2
## <chr> <dbl> <dbl>
## 1 gene1 3 4
## 2 gene2 5 6这个是最基本的长宽格式数据转换,如果数据集有很多列,有时候转换会相对比较复杂,这里就不做介绍,因为我也搞不懂有时候
8.2 ggplot2热图
以下介绍ggplot2做热图的代码都是假设已经拿到了长格式数据
示例数据如下
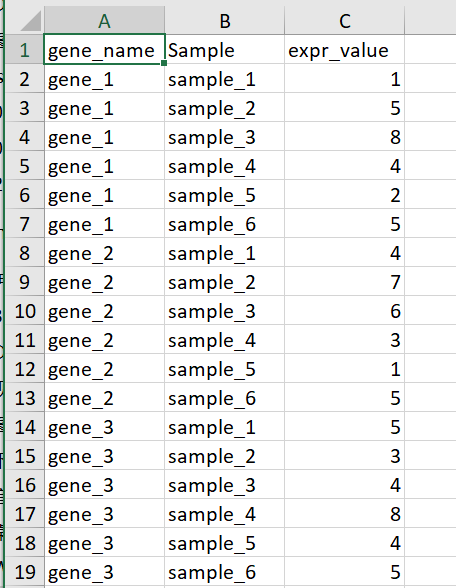
最基本的热图代码
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5library(ggplot2)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")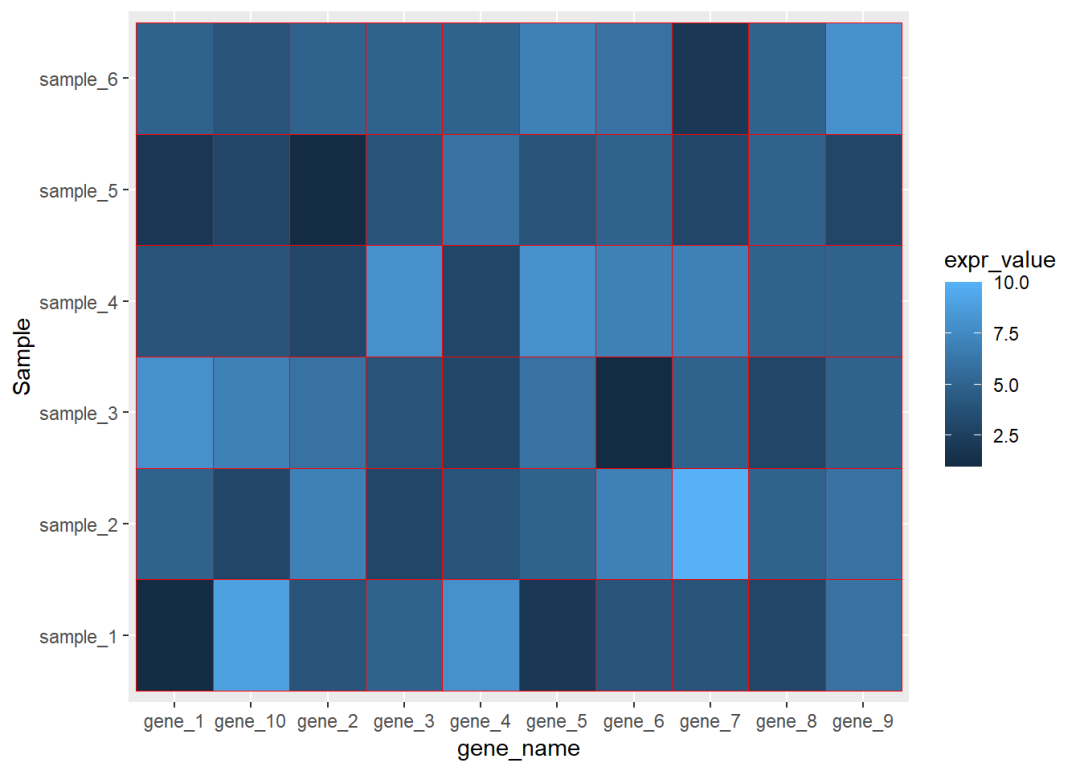
ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color=NA)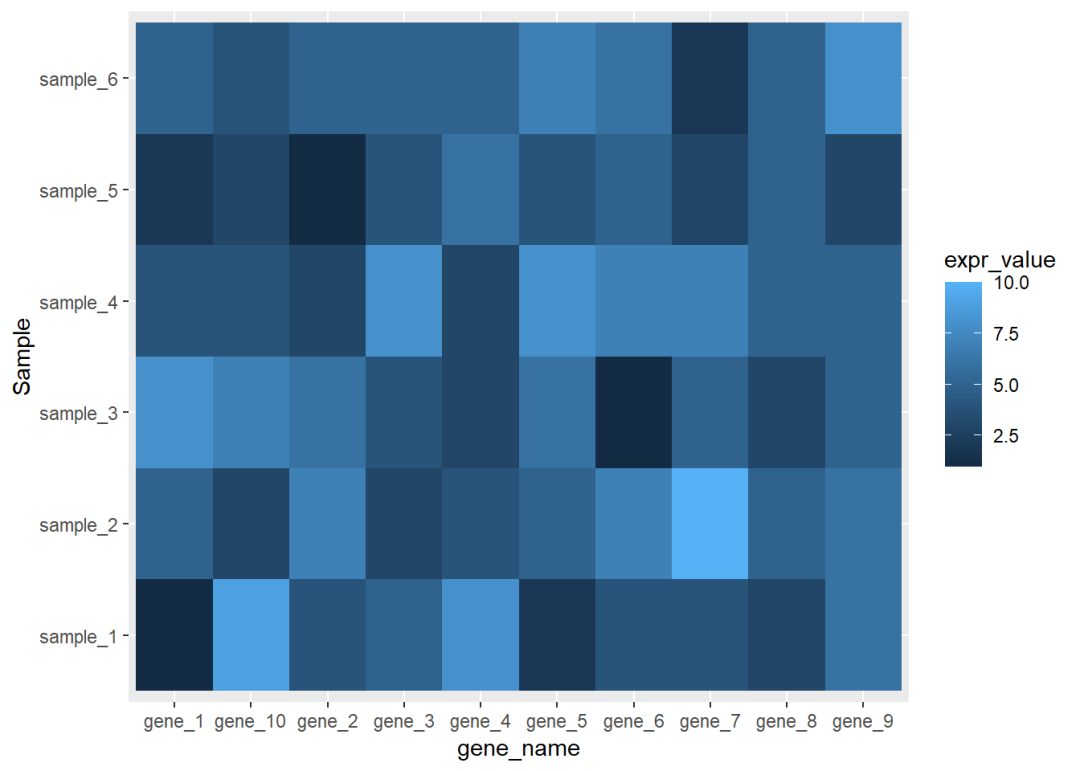
热图经常遇到的操作是调整坐标轴的顺序,这个可以通过赋予因子水平来实现
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")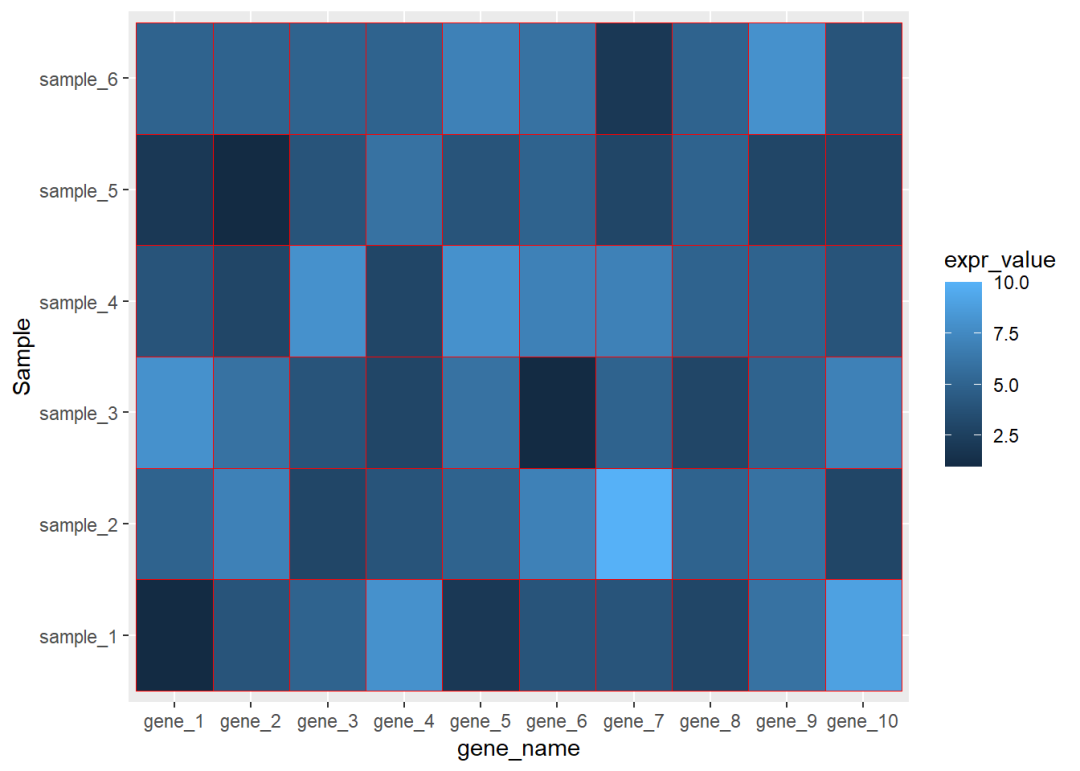
dat01$Sample<-factor(dat01$Sample, levels = c("sample_2","sample_4","sample_6", "sample_1","sample_3","sample_5"))ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")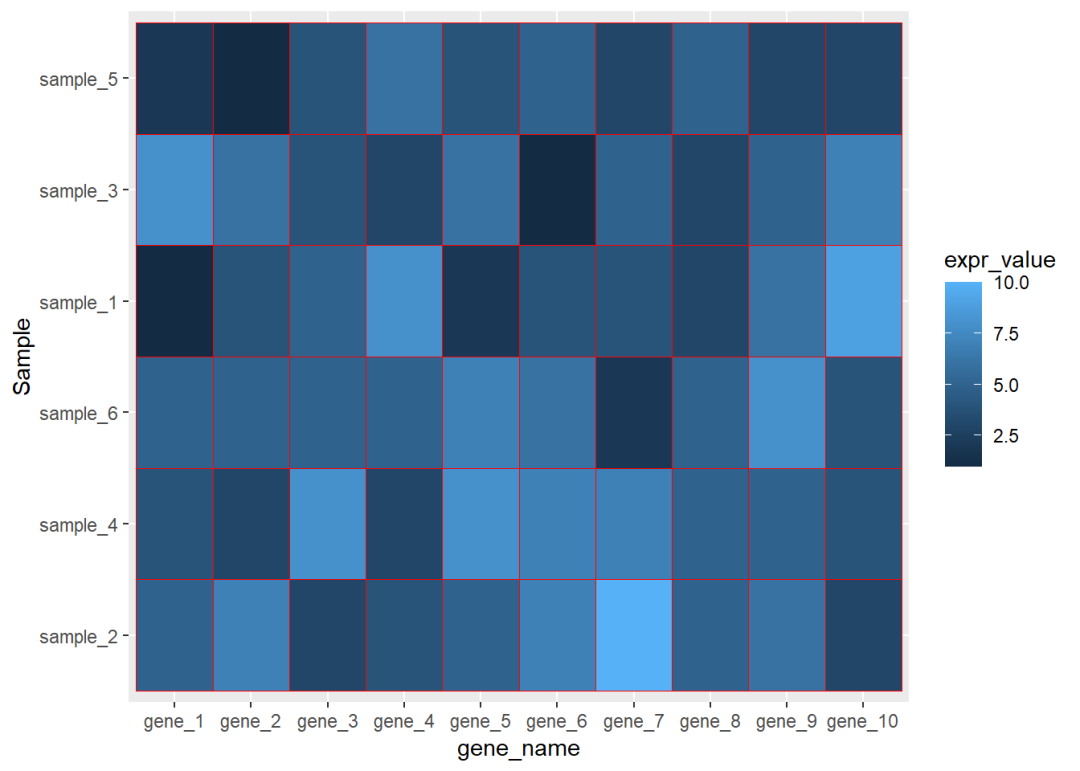
更改热图色块填充的颜色
更改热图填充颜色有很多种方式,这里我介绍我自己最常用的一种方式
参考链接
https://r-charts.com/
这里用到额外的一个R包 paletteer
https://github.com/EmilHvitfeldt/paletteer
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")
library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue")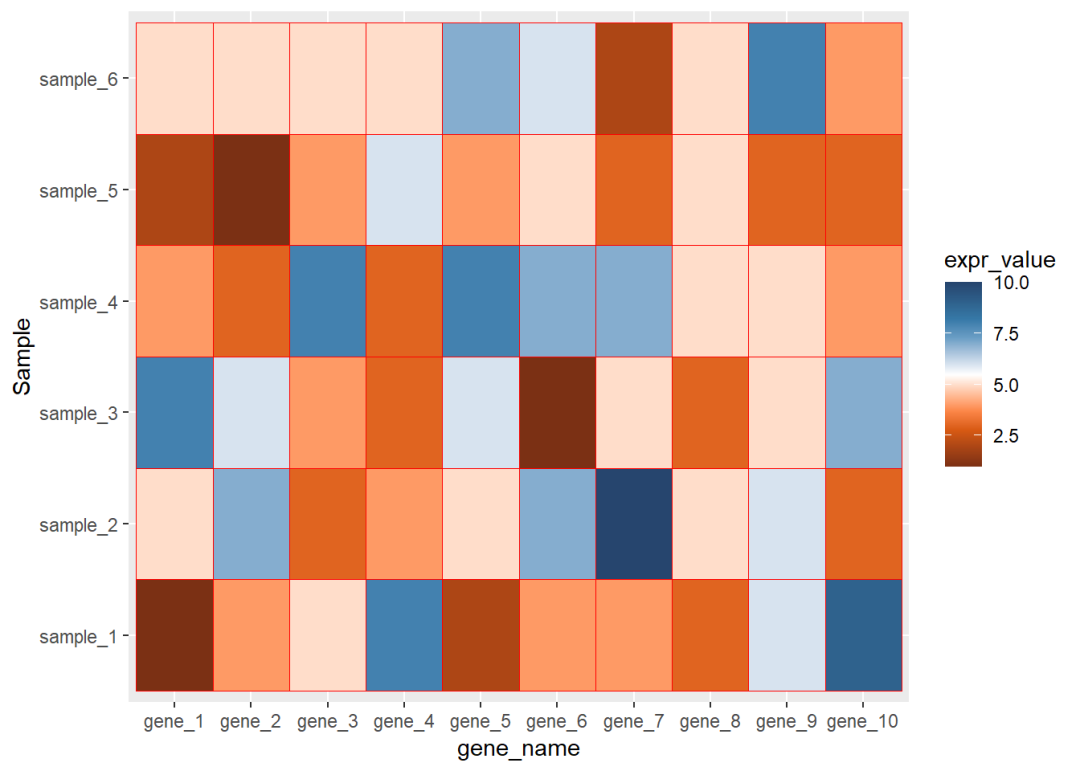
ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)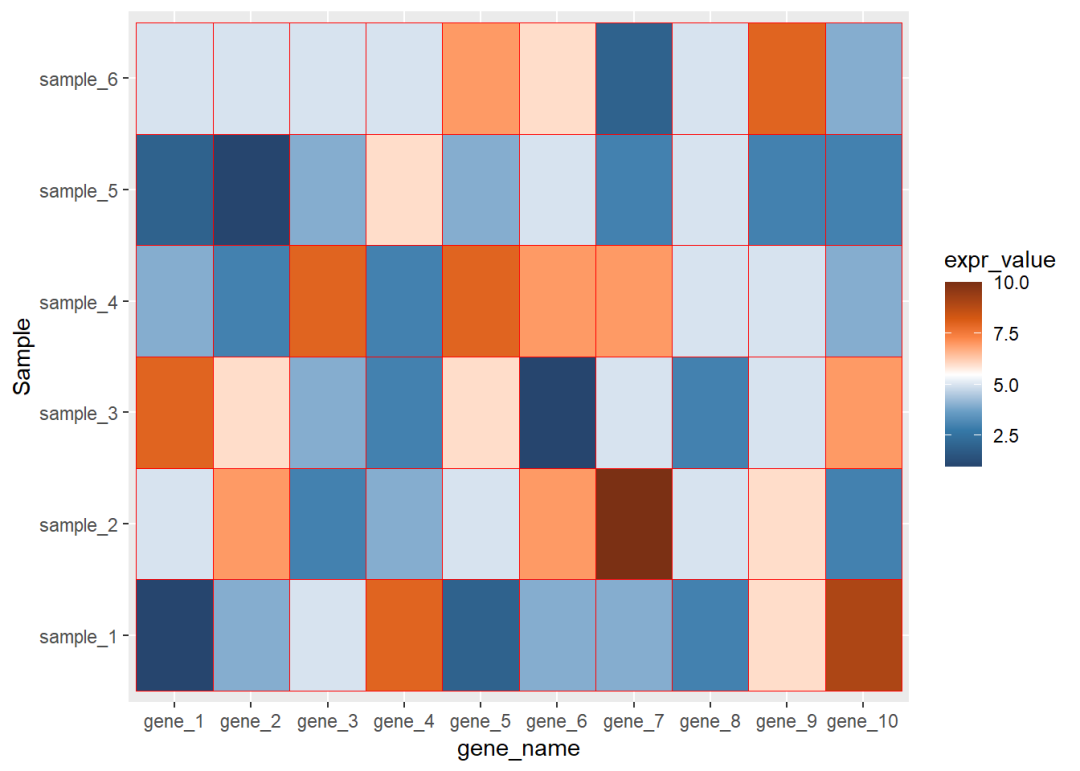
调整坐标轴文本标签的位置,y轴左右,x轴是上下
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)+ scale_x_discrete(position = "top")+ scale_y_discrete(position = "right")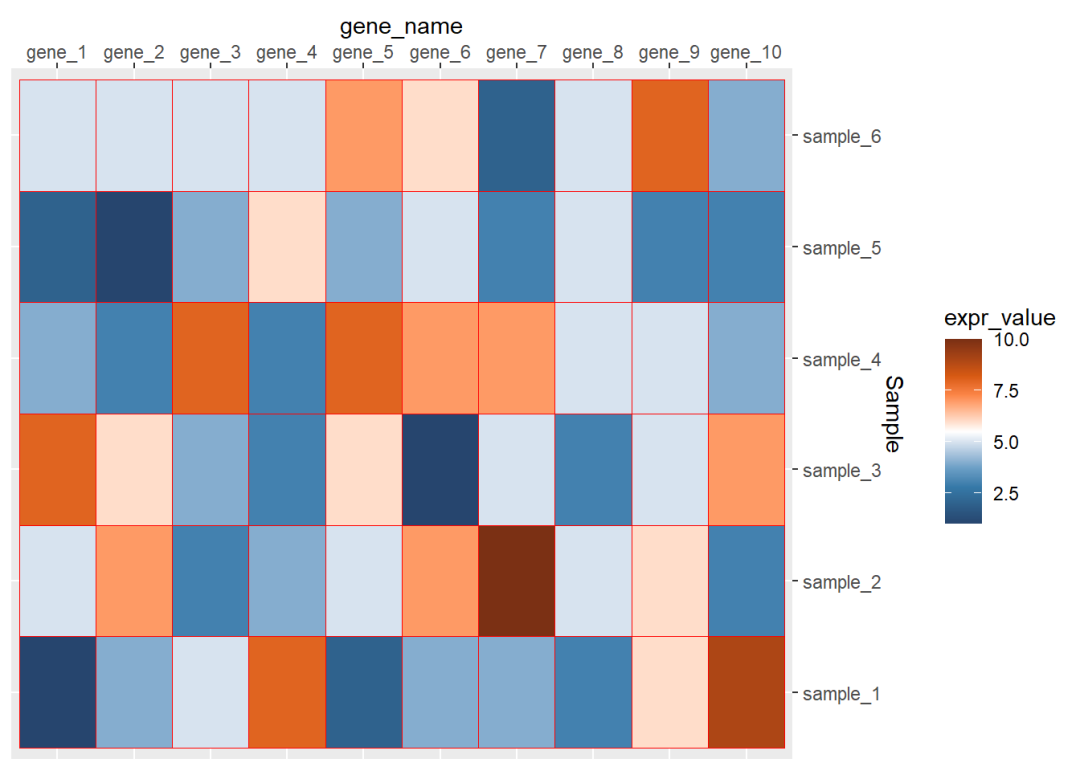
调整坐标轴的文本方向
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)+ scale_x_discrete(position = "top")+ scale_y_discrete(position = "right")+ theme(axis.text.x = element_text(angle=60))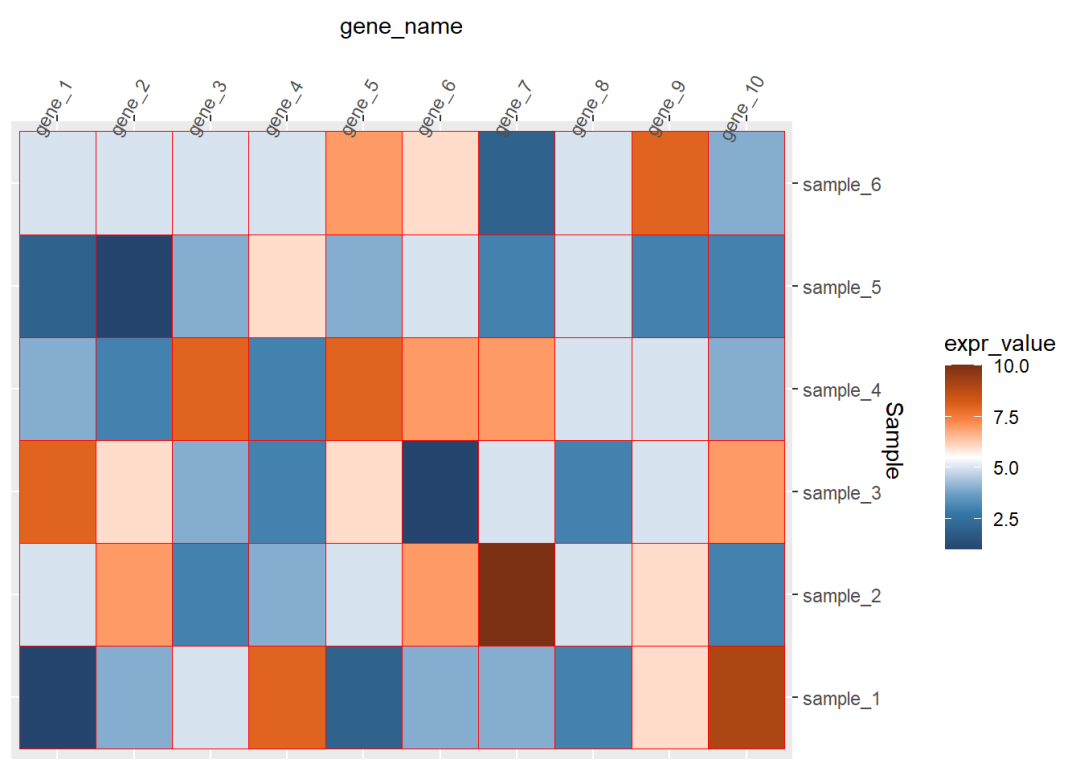
ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)+ scale_x_discrete(position = "top")+ scale_y_discrete(position = "right")+ theme(axis.text.x = element_text(angle=60,hjust=0,vjust=1))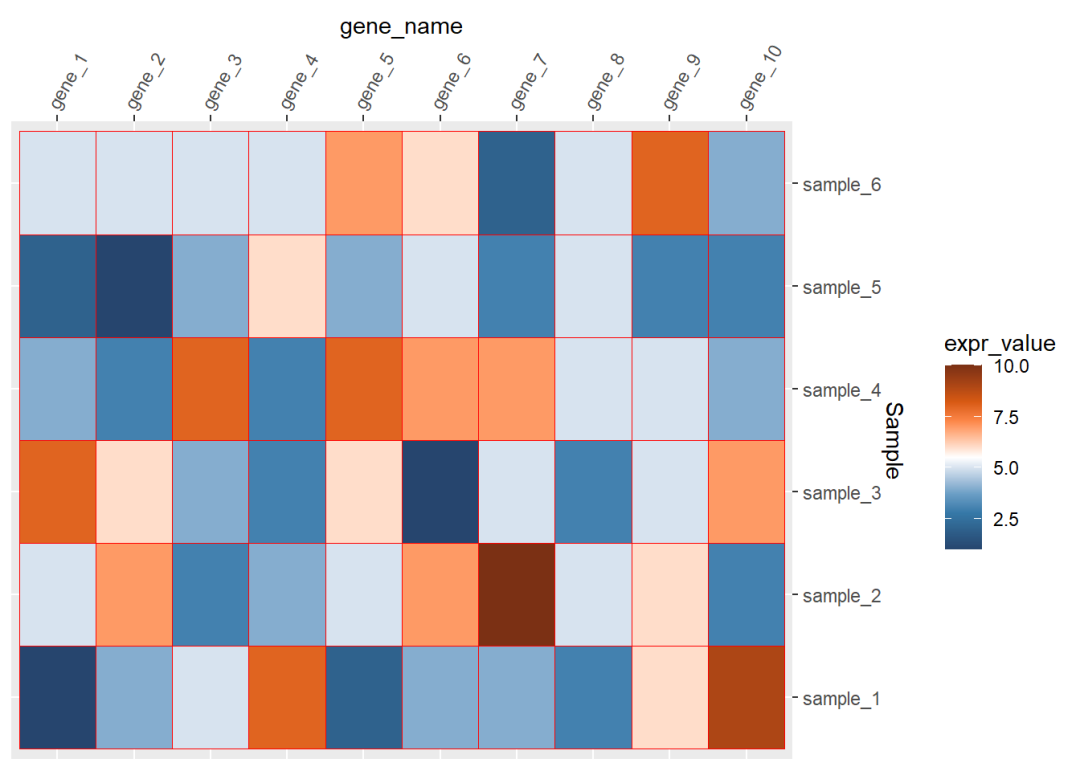
去掉整个的灰色背景和坐标轴的小短线
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)+ scale_x_discrete(position = "top")+ scale_y_discrete(position = "right")+ theme(axis.text.x = element_text(angle=60,hjust=0,vjust=1), axis.ticks = element_blank(), panel.background = element_blank())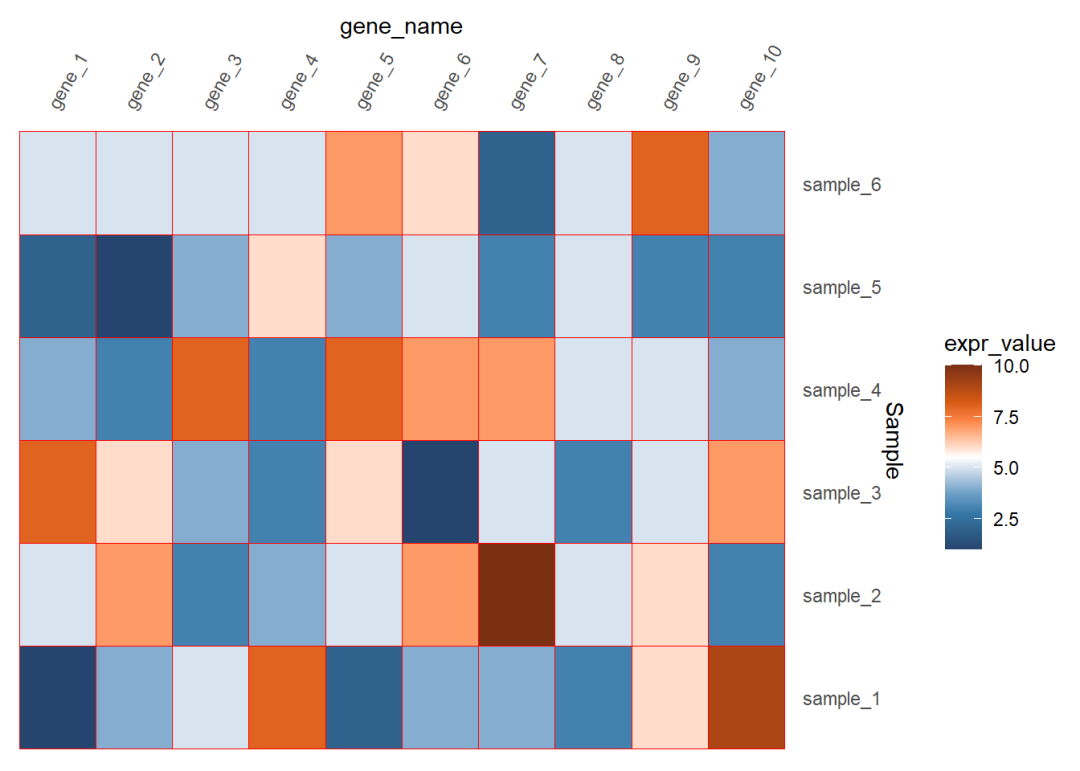
如果看起来坐标轴的文本距离图比较远的话还可以调整
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)+ scale_x_discrete(position = "top", expand = expansion(mult = c(0,0)))+ scale_y_discrete(position = "right", expand = expansion(mult = c(0,0)))+ theme(axis.text.x = element_text(angle=60,hjust=0,vjust=1), axis.ticks = element_blank(), panel.background = element_blank())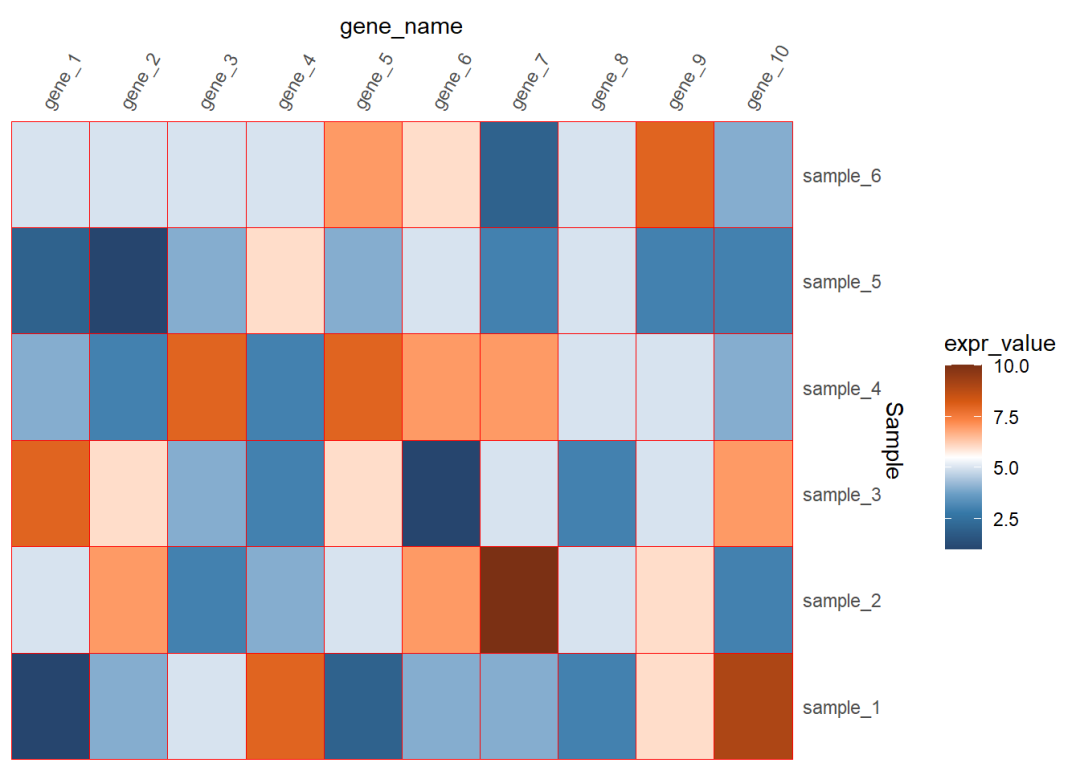
在色块上添加文本,用geom_text()函数来添加
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)+ scale_x_discrete(position = "top", expand = expansion(mult = c(0,0)))+ scale_y_discrete(position = "right", expand = expansion(mult = c(0,0)))+ theme(axis.text.x = element_text(angle=60,hjust=0,vjust=1), axis.ticks = element_blank(), panel.background = element_blank())+ geom_text(aes(label=expr_value))调整图例的细节
参考公众号推文 ggplot2画热图展示相关系数的简单小例子
截断和标签是在scale_fill函数里设置breaks和labels
图例的位置是在主题里进行设置
其他一些细节在guides函数里设置
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr_value),color="red")+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1, breaks=c(1.1,3,5,7,9,9.9), labels=c(1,3,5,7,9,10))+ scale_x_discrete(position = "top", expand = expansion(mult = c(0,0)))+ scale_y_discrete(position = "right", expand = expansion(mult = c(0,0)))+ theme(axis.text.x = element_text(angle=60,hjust=0,vjust=1), axis.ticks = element_blank(), panel.background = element_blank())+ geom_text(aes(label=expr_value))+ theme(legend.position = "top")+ guides(fill=guide_colorbar(title = "AAA", title.position = "top", title.hjust = 0.5, barwidth = 20, ticks = FALSE, label = TRUE))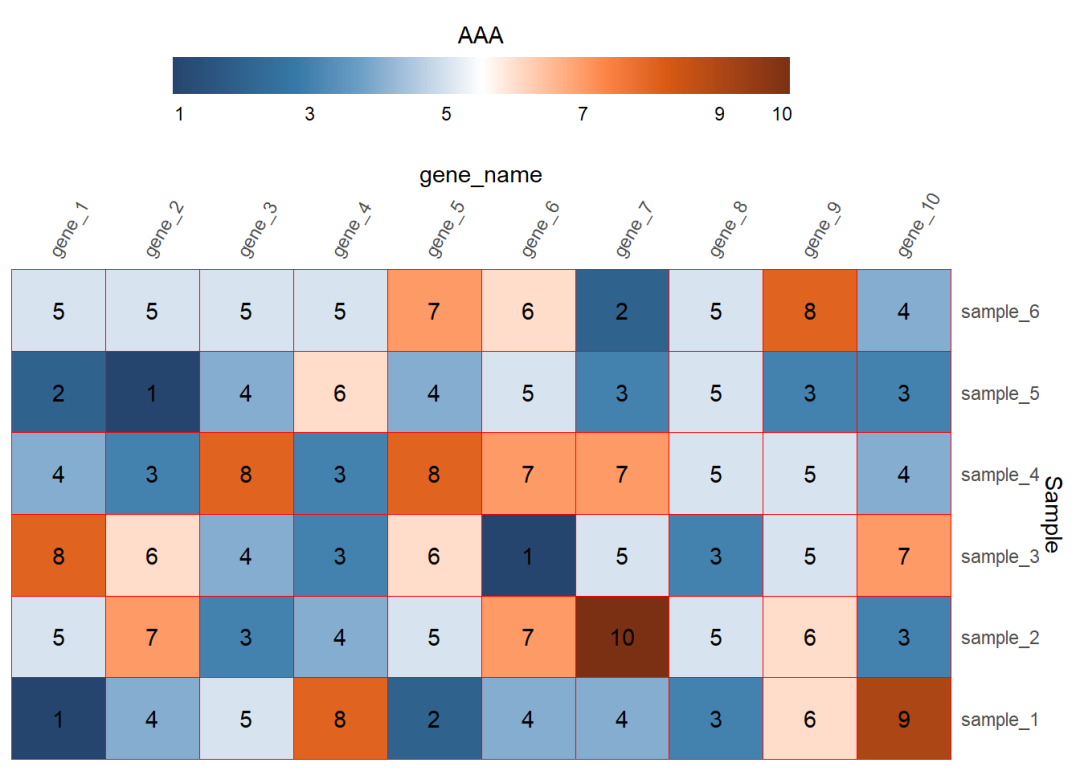
以上介绍的用来填充颜色的数据是连续型的,数据是离散的也是可以的,比如只关心某个基因在样本中是否表达,并不关心这个基因的表达量高低,示例数据如下
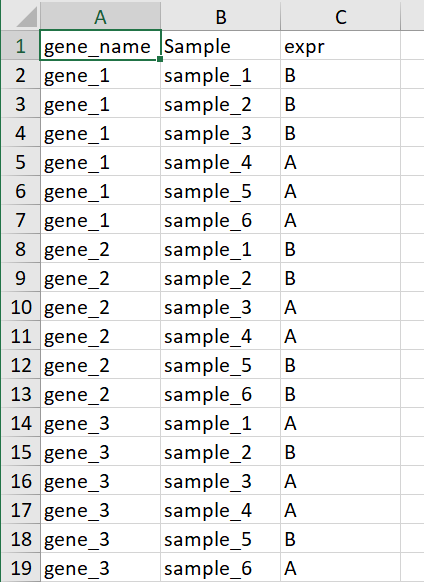
这里A代表基因表达B代表基因不表达,这个AB可以用任意字符代替
library(readxl)dat01<-read_excel("example_data/08-heatmap/04_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr
## <chr> <chr> <chr>
## 1 gene_1 sample_1 B
## 2 gene_1 sample_2 B
## 3 gene_1 sample_3 B
## 4 gene_1 sample_4 A
## 5 gene_1 sample_5 A
## 6 gene_1 sample_6 Adat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr),color="blue")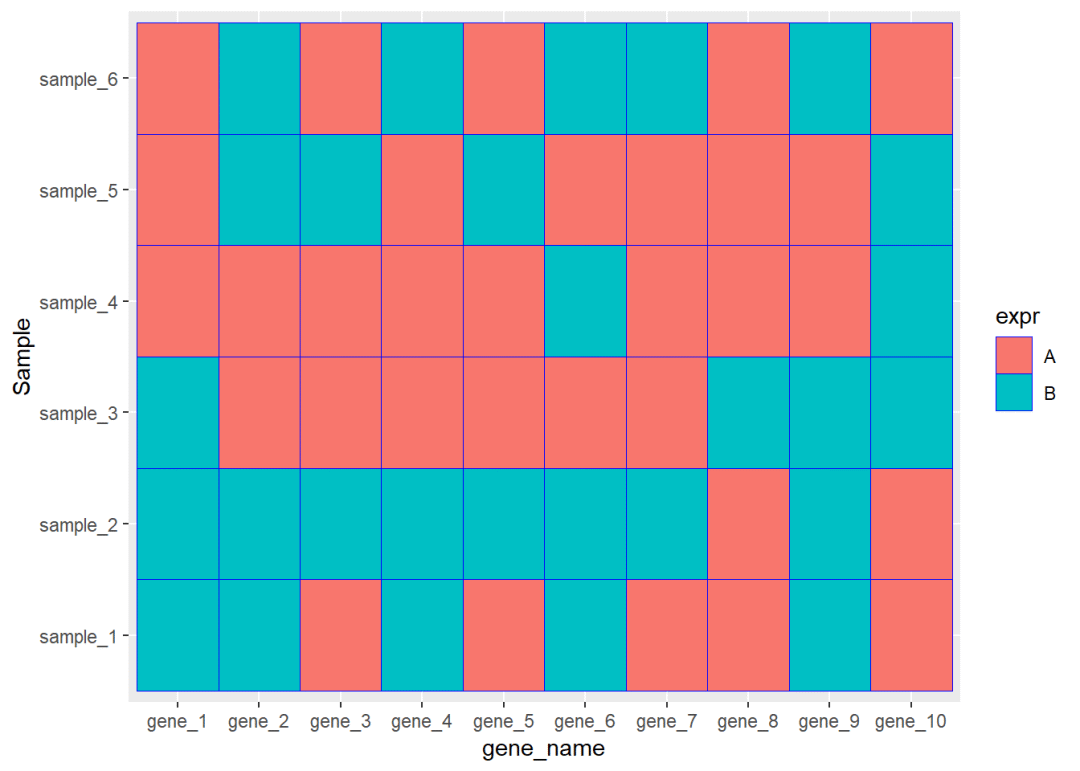
ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr),color="blue")+ scale_fill_manual(values = c("A"="#fe0904", "B"='#f9b54f'))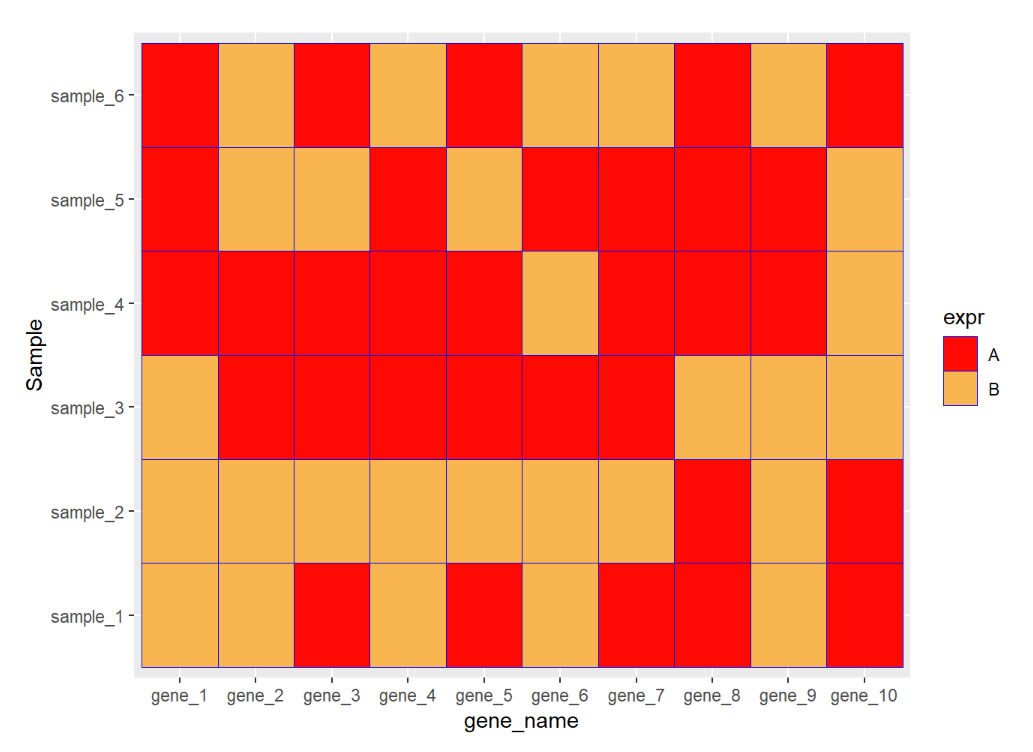
ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_tile(aes(fill=expr),color="blue")+ scale_fill_manual(values = c("A"="#fe0904", "B"='white'))+ theme_bw()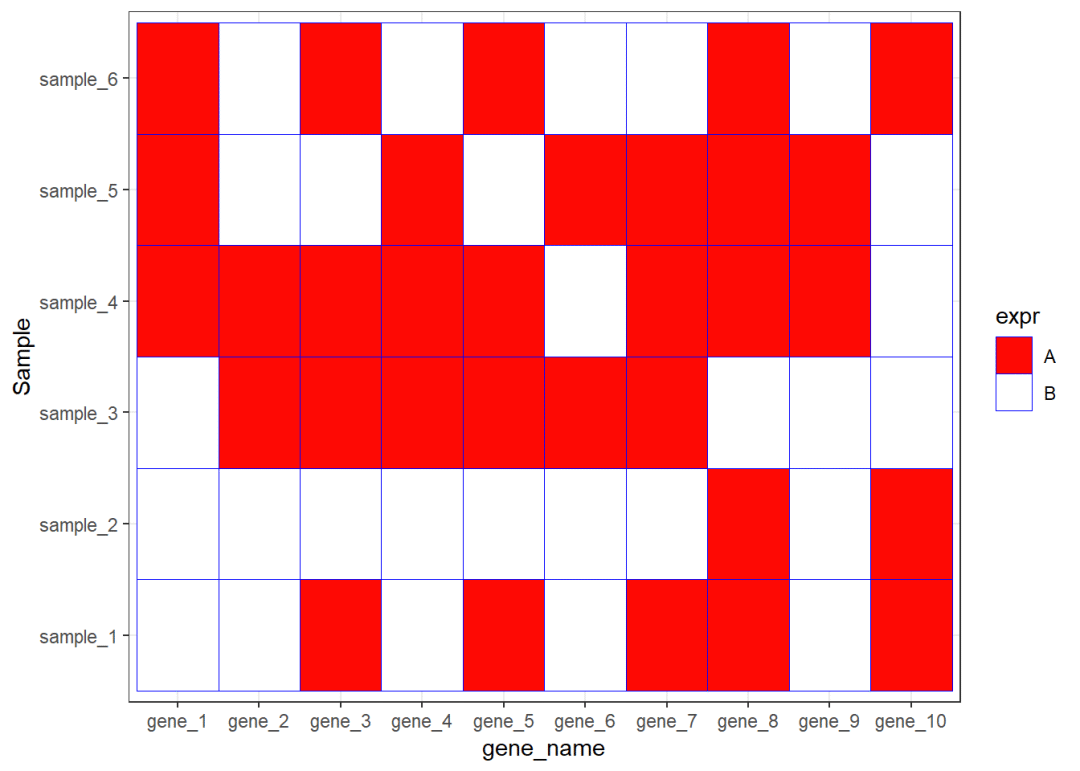
8.3 ggplot2气泡热图图
如果x 和 y都是离散的,把热图函数geom_tile()换成geom_point()函数,然后用表达量的值映射点的大小 同时映射颜色 也可以归为热图的一种
比如
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_point(aes(fill=expr_value, size=expr_value),color="blue",shape=21)
更改默认的配色和点的大小
library(readxl)dat01<-read_excel("example_data/08-heatmap/03_heatmap_example.xlsx")head(dat01)## # A tibble: 6 x 3
## gene_name Sample expr_value
## <chr> <chr> <dbl>
## 1 gene_1 sample_1 1
## 2 gene_1 sample_2 5
## 3 gene_1 sample_3 8
## 4 gene_1 sample_4 4
## 5 gene_1 sample_5 2
## 6 gene_1 sample_6 5dat01$gene_name<-factor(dat01$gene_name, levels = c("gene_1","gene_2","gene_3", "gene_4","gene_5","gene_6", "gene_7","gene_8","gene_9", "gene_10"))library(ggplot2)library(paletteer)ggplot(data=dat01,aes(x=gene_name,y=Sample))+ geom_point(aes(fill=expr_value, size=expr_value),color="blue",shape=21)+ scale_fill_paletteer_c("ggthemes::Classic Orange-White-Blue", direction = -1)+ scale_size_continuous(range = c(1,20), guide = guide_legend(override.aes = list(size = c(1,2,3,5)) ))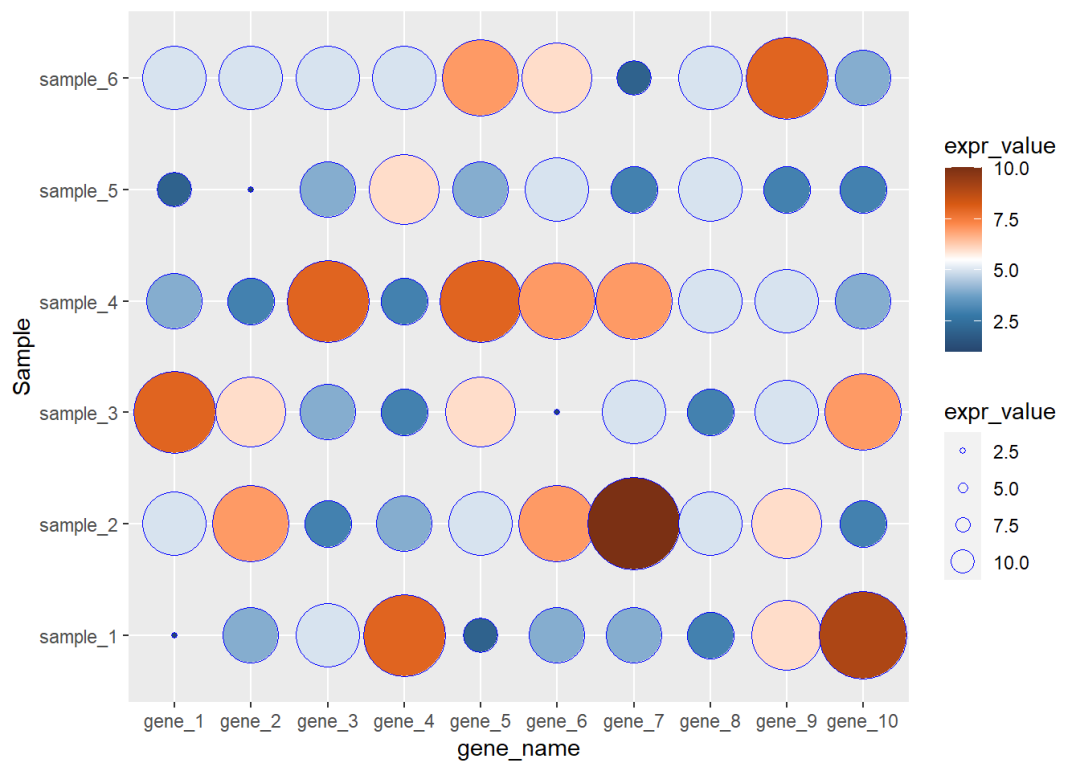
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2022-06-11,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

