monocle3轨迹分析
原创
https://mp.weixin.qq.com/s/UsDC-t1j7NHaLTnI6xCATQ
monocle3与PAGA有点类似,在UMAP图上显示轨迹图,没有了树状的结构。
原理、图的理解,可以参考Reference中的链接
安装
- ubuntu
sudo apt install libudunits2-dev libgdal-dev- R
speedglm包违反CRAN规定被删除了,所以要从rstudio下载
using(pak)
pkgs <- c('BiocGenerics', 'DelayedArray', 'DelayedMatrixStats',
'limma', 'lme4', 'S4Vectors', 'SingleCellExperiment',
'SummarizedExperiment', 'batchelor', 'HDF5Array',
'terra', 'ggrastr',"hhoeflin/hdf5r","mojaveazure/loomR@develop",
"url::https://packagemanager.rstudio.com/cran/2023-03-31/src/contrib/speedglm_0.3-4.tar.gz")
pak::pkg_install(pkgs)0. 加载R包、定义函数
using(monocle3, tidyverse, magrittr, Matrix, Seurat, SeuratObject, sp)
ps <- function(filename, plot = FALSE, w = 9, h = 6) {
if (is.object(plot)) {
print(plot)
}
plot <- recordPlot()
pdf(file = paste0(filename,".pdf"), onefile = T, width = w, height = h)
replayPlot(plot)
dev.off()
}
n_jobs <- 8 # 使用的线程数1.创建monocle对象
- sobj:Seurat对象
- cell_type:已经注释好了细胞类型
- orig.ident:批次信息
- sobj_embed:UMAP降维信息,是数据框,行名是细胞,有两列分别对应两个维度
- hvg:3000个高变基因用于后续降维
expression_matrix, a numeric matrix of expression values, where rows are genes, and columns are cellscell_metadata, a data frame, where rows are cells, and columns are cell attributes (such as cell type, culture condition, day captured, etc.)gene_metadata, an data frame, where rows are features (e.g. genes), and columns are gene attributes, such as biotype, gc content, etc. one of the columns of thegene_metadatashould be named "gene_short_name", which represents the gene symbol or simple name (generally used for plotting) for each gene.
sobj <- readRDS("final.rds")
gene_metadata <- sobj@assays$RNA@meta.features
gene_metadata$gene_short_name <- rownames(gene_metadata)
cds <- new_cell_data_set(as(sobj@assays$RNA@counts,"sparseMatrix"),
cell_metadata = sobj@meta.data,
gene_metadata = gene_metadata
)
hvg <- seob@assays$integrated@var.features
sobj_embed <- Seurat::Embeddings(seob, reduction = "umap")
rm(sobj, gene_metadata)2.pre-process
标准化、预处理、取批次
## Normalize and pre-process the data
cds %<>% preprocess_cds(num_dim = 30,method="PCA",norm_method="log",use_genes=hvg)
## Remove batch effects with cell alignment
cds %<>% align_cds(alignment_group = "orig.ident",preprocess_method="PCA")3.Reduce dimensions and Cluster cells
降维、聚类、分群、分partition
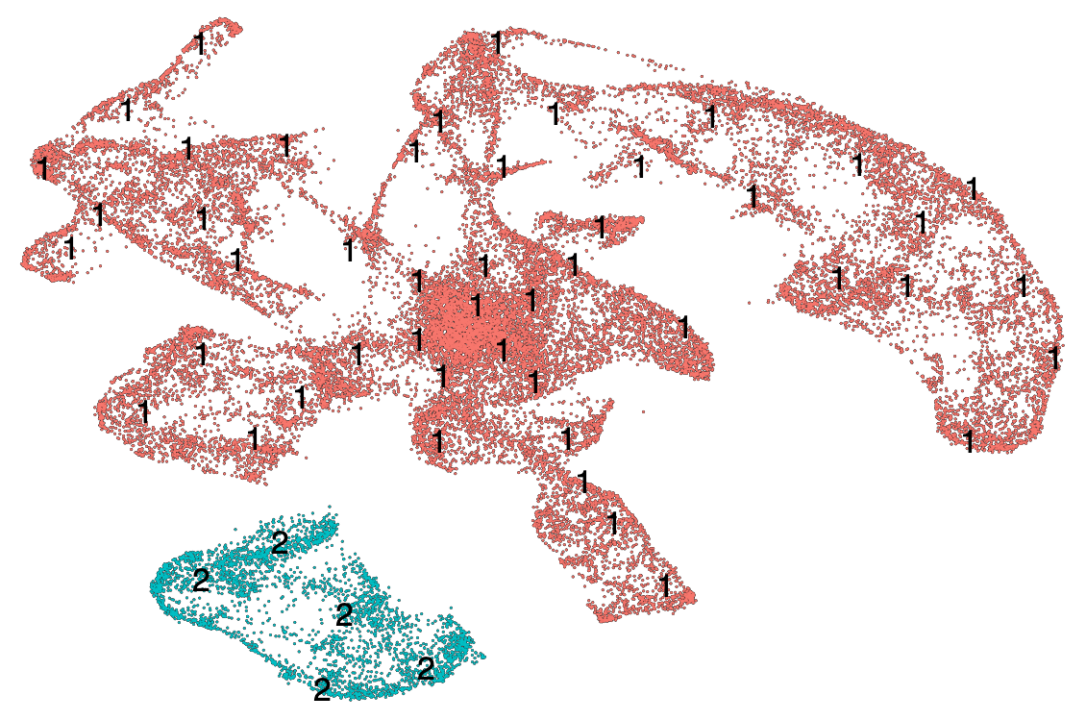
这里使用UMAP作为降维算法,再使用轨迹分区算法,把所有细胞分为两个partitio,不同分区的细胞会进行单独的轨迹分析。
plot_pc_variance_explained(cds)
ps("PCA.pdf", w = 9, h = 6)
## Reduce the dimensions using UMAP
cds %<>% reduce_dimension(umap.fast_sgd = TRUE, cores = n_jobs, reduction_method = "UMAP", preprocess_method = "PCA")
## 从seurat导入整合过的umap坐标
cds_embed <- cds@int_colData$reducedDims$UMAP
cds@int_colData$reducedDims$UMAP <- sobj_embed[rownames(cds_embed),]
plot_cells(cds, reduction_method = "UMAP", color_cells_by = "cell_type",group_label_size=6)
ps("UMAP_CellType.pdf")
## Cluster the cells
cds %<>% rcluster_cells(reduction_method = "UMAP", cluster_method = "leiden",random_seed=1314)
plot_cells(cds, color_cells_by = "partition",group_label_size=6)
ps("UMAP_partition.pdf")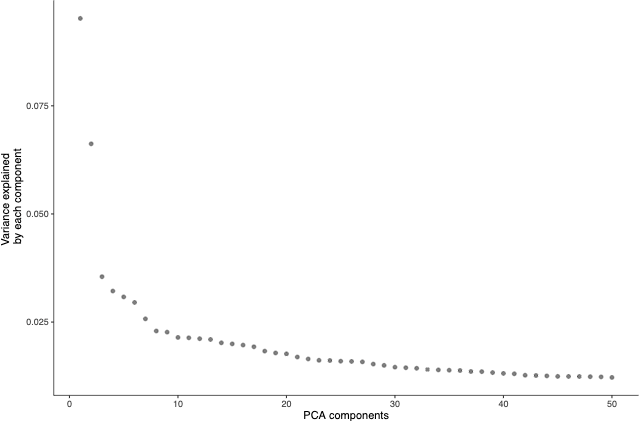
4.Order cells in pseudotime along a trajectory
手动选择root需要根据自己的生物学背景知识
## Learn a graph
cds %<>% learn_graph()
plot_cells(cds,trajectory_graph_segment_size = 1.5,
group_label_size = 6,group_cells_by="cluster",
color_cells_by = "cell_type", label_groups_by_cluster = FALSE,
label_leaves = FALSE, label_branch_points = FALSE
)
ps("UMAP_graph.pdf")
cds <- order_cells(cds)
plot_cells(cds,
group_label_size = 6,
color_cells_by = "pseudotime",
label_cell_groups = FALSE,
label_leaves = FALSE,
label_branch_points = FALSE,
graph_label_size = 1.5
)
ps("Trajectory_Pseudotime.pdf")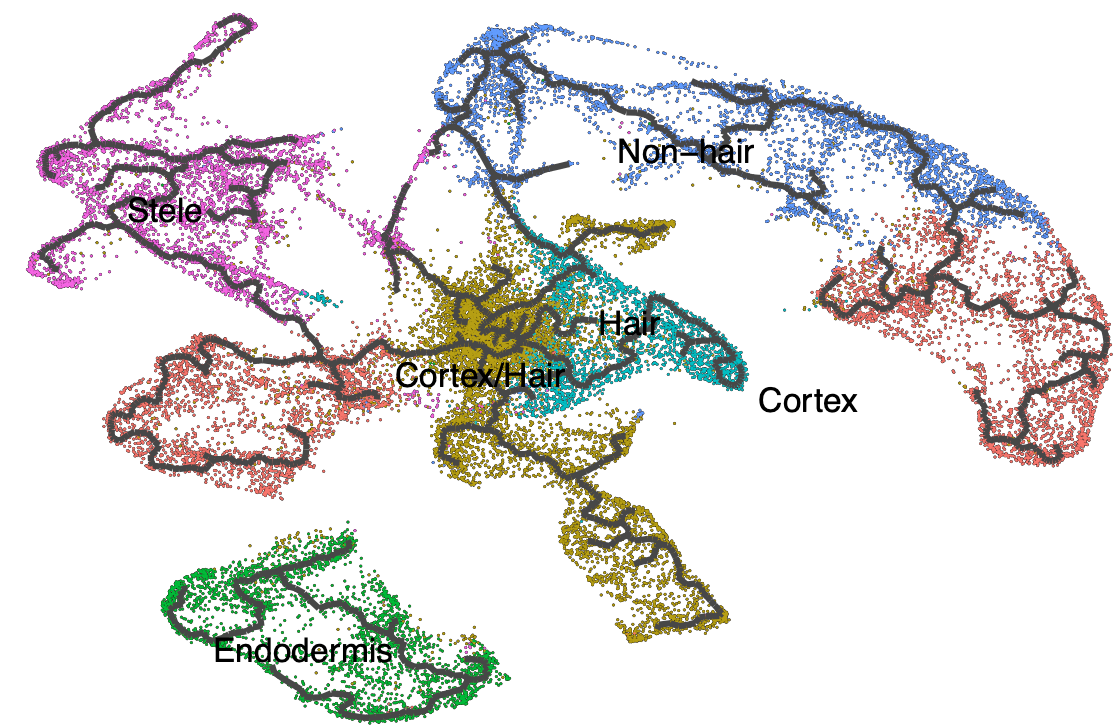
The black lines show the structure of the graph.
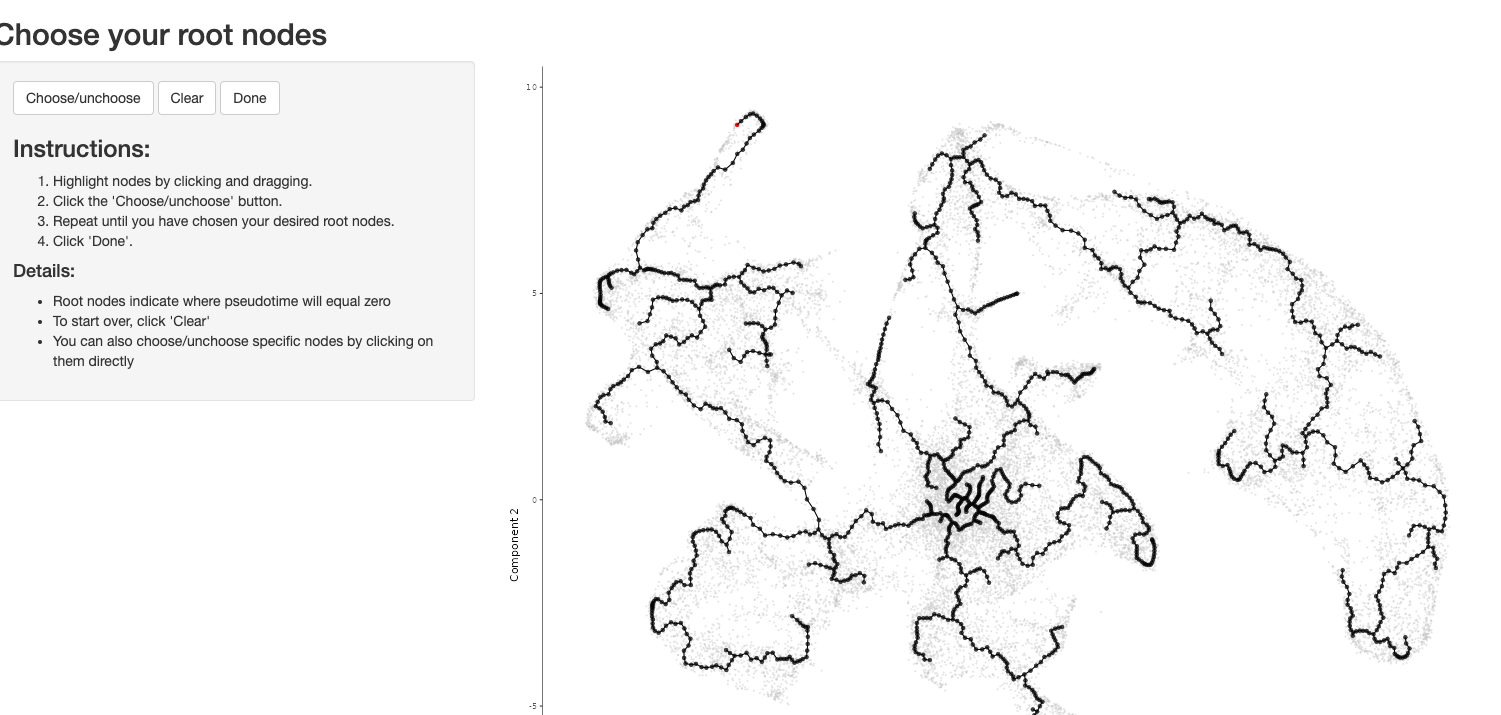
手动选择root
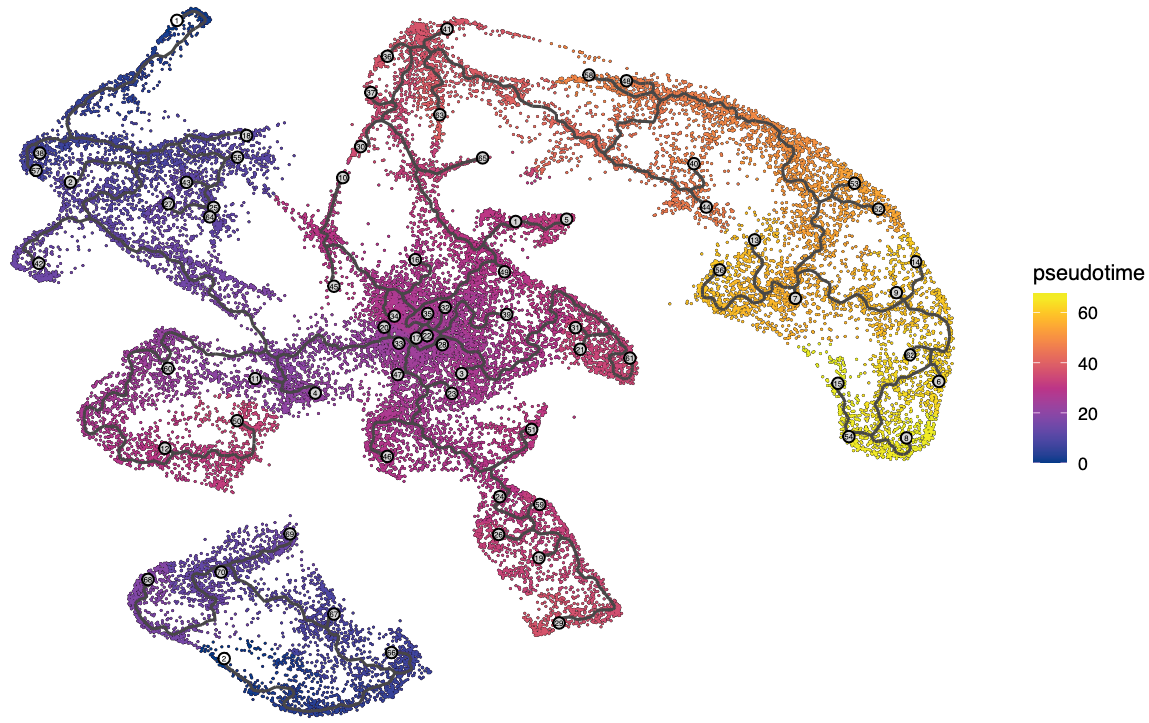
The black lines show the structure of the graph. Note that the graph is not fully connected: cells in different partitions are in distinct components of the graph. The circles with numbers in them denote special points within the graph. Each leaf, denoted by light gray circles, corresponds to a different outcome (i.e. cell fate) of the trajectory. Black circles indicate branch nodes, in which cells can travel to one of several outcomes.
5.Perform differential expression analysis
这是空间差异分析常用的方法,在空间转录组中也有。
morans_Icolumn, which ranges from -1 to +1. A value of 0 indicates no effect, while +1 indicates perfect positive autocorrelation and suggests that nearby cells have very similar values of a gene's expression. Significant values much less than zero are generally rare.
dea_res <- graph_test(cds, neighbor_graph = "principal_graph", cores = n_jobs) %>%
dplyr::filter(q_value < 0.05)
fwrite(dea_res, "Trajectory_genes.csv")6.Finding genes that change as a function of pseudotime
寻找拟时轨迹差异基因,使用graph_test分析最重要的结果是莫兰指数(morans_I),其值在-1至1之间,0代表此基因没有。空间共表达效应,1代表此基因在空间距离相近的细胞中表达值高度相似。根据莫兰指数挑选前10个基因用于可视化。
degs <- dea_res$gene_short_name
top_genes <- dea_res %>%
dplyr::top_n(n = 10, morans_I) %>%
dplyr::pull(gene_short_name) %>%
as.character()
plot_genes_in_pseudotime(cds[top_genes, ],
color_cells_by = "cell_type",
min_expr = 0.5, ncol = 2
)
ps("Top_Genes_Jitterplot.pdf", w = 9, h = 6)
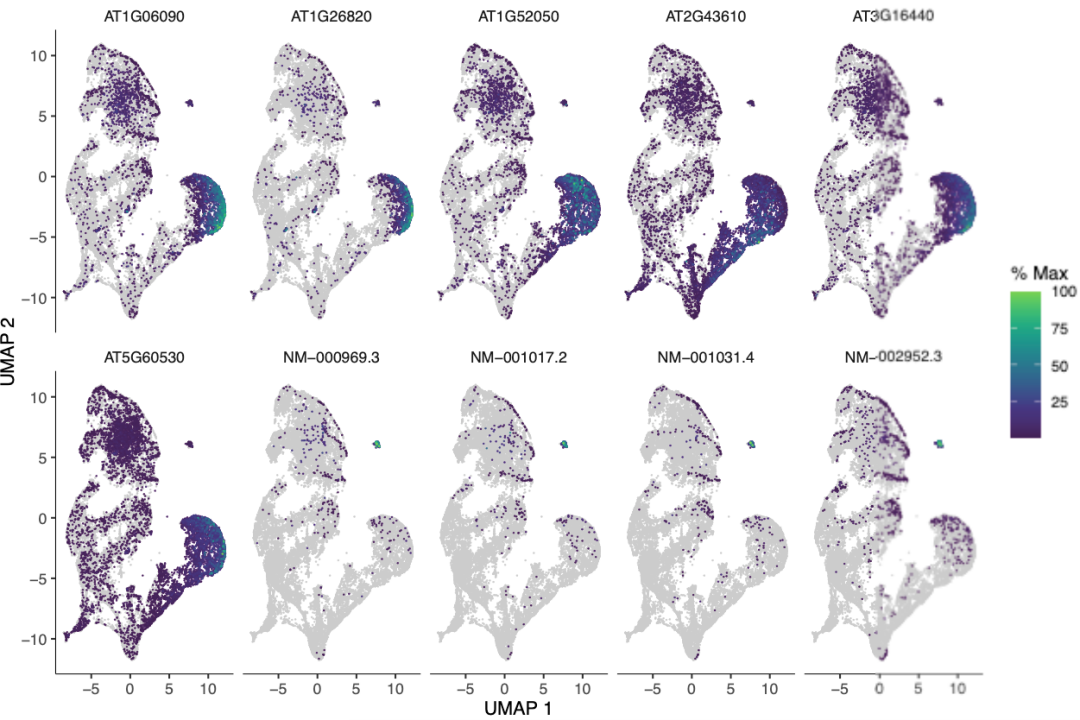
p <- plot_cells(cds,
genes = top_genes, show_trajectory_graph = FALSE,
label_cell_groups = FALSE, label_leaves = FALSE
)
p$facet$params$ncol <- 5
ps("Top_Genes_Featureplot.pdf", plot = p)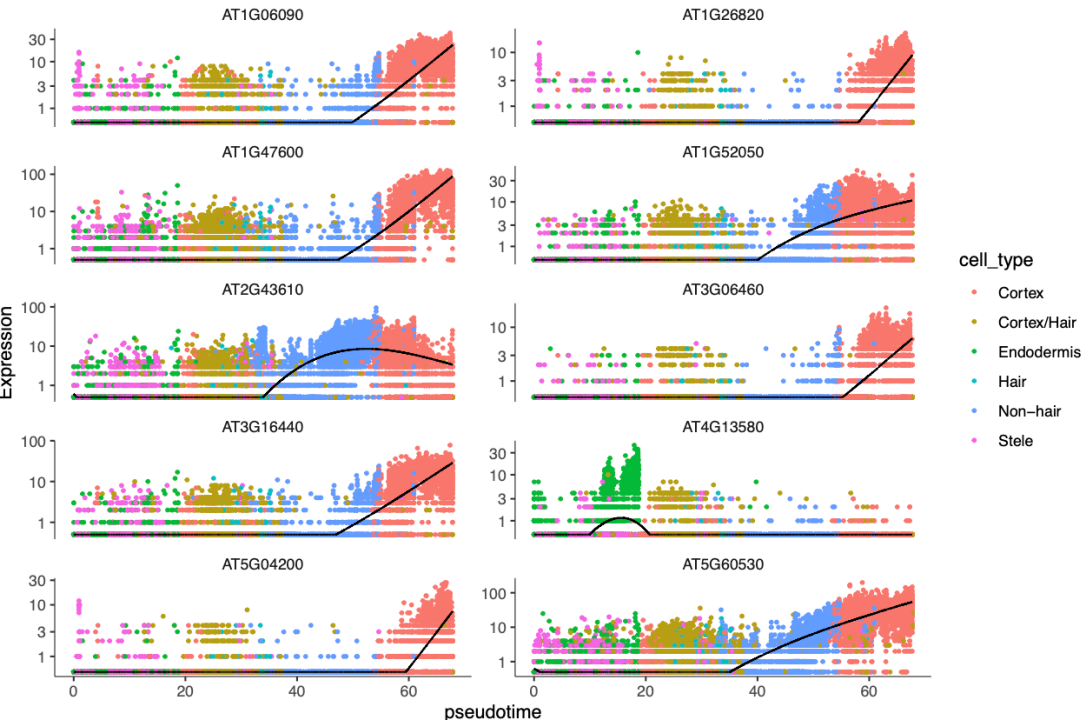
挑选top10画图展示,基因表达随时间变化趋势图
7.Finding modules of co-regulated genes
寻找共表达基因模块,根据上边的差异分析结果,按照UMAP和Louvain 聚类,将这些基因分在不同的模块中,有些模块在某些细胞中特异高表达。
Once we have a set of genes that vary in some interesting way across the clusters, Monocle provides a means of grouping them into modules. We can call find_gene_modules(), which essentially runs UMAP on the genes (as opposed to the cells) and then groups them into modules using Louvain community analysis
gene_module_df <- find_gene_modules(cds[degs, ], resolution = 1e-2, cores = n_jobs)
fwrite(gene_module_df, "Genes_Module.csv")
cell_group_df <- tibble::tibble(
cell = row.names(colData(cds)),
cell_group = colData(cds)$cell_type
)
agg_mat <- aggregate_gene_expression(cds, gene_module_df, cell_group_df)
row.names(agg_mat) <- stringr::str_c("Module ", row.names(agg_mat))
pheatmap::pheatmap(agg_mat,
cluster_rows = TRUE, cluster_cols = TRUE,
scale = "column", clustering_method = "ward.D2",
fontsize = 6
)
ps("heatmap.pdf", w = 5, h = 16)
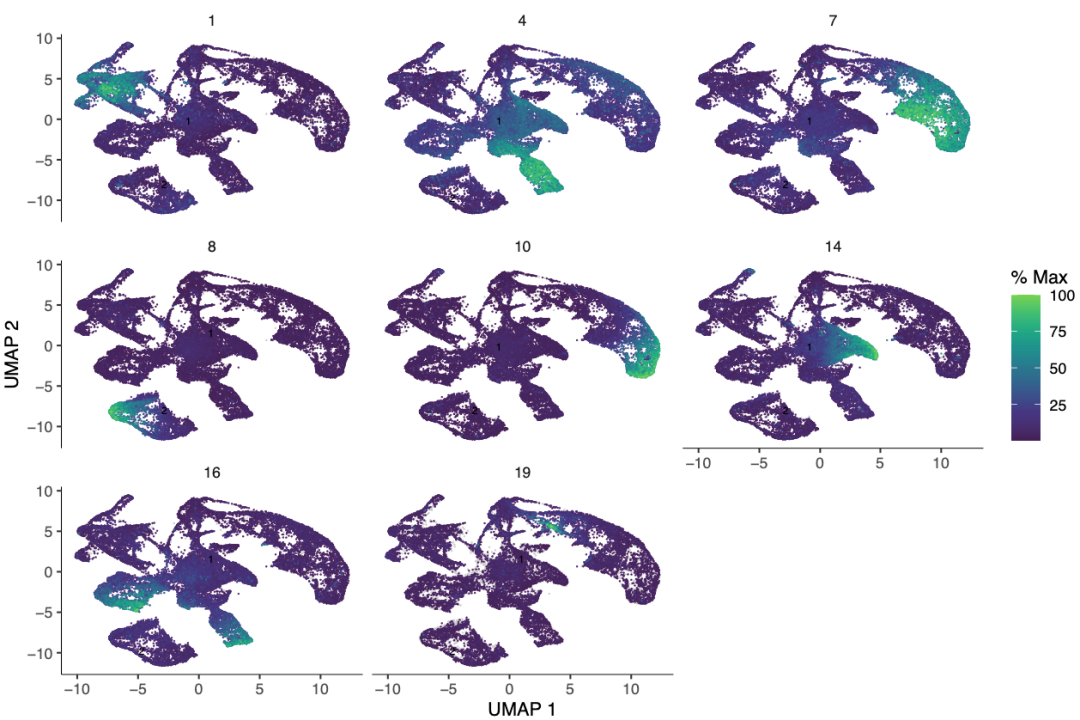
plot_cells(cds,
genes = gene_module_df %>% dplyr::filter(module %in% c(200, 169, 138, 22,17,91)),
group_cells_by = "partition",
color_cells_by = "partition",
show_trajectory_graph = FALSE
)
ps("module.pdf")
saveRDS(cds,file="cds.rds")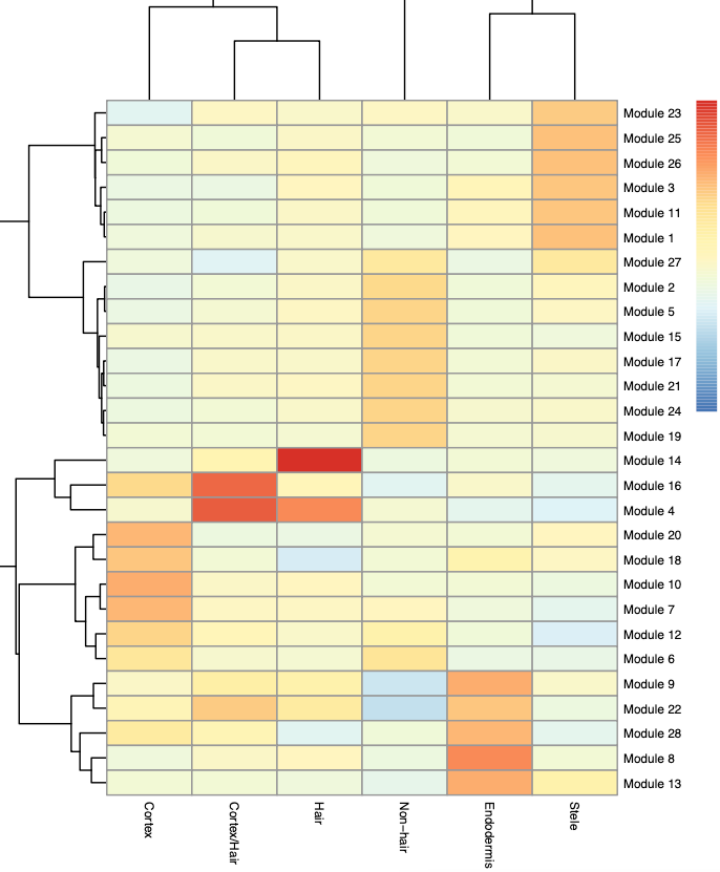
Reference
https://zhuanlan.zhihu.com/p/451727080
https://cole-trapnell-lab.github.io/monocle3/docs
https://www.jianshu.com/p/c402b6588e17原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录


