单细胞韧皮部研究代码解析4-Fig01-umap_cell_types.R
原创单细胞韧皮部研究代码解析4-Fig01-umap_cell_types.R
原创
小胡子刺猬的生信学习123
发布于 2023-05-06 17:20:34
发布于 2023-05-06 17:20:34
单细胞韧皮部研究代码解析1-QC_filtering.R:https://cloud.tencent.com/developer/article/2256814?areaSource=&traceId=
单细胞韧皮部研究代码解析2--comparison_denyer2019.R:https://cloud.tencent.com/developer/beta/article/2260381?areaSource=&traceId=
单细胞韧皮部研究代码解析3-comparison_brady.R:https://cloud.tencent.com/developer/beta/article/2280058?areaSource=&traceId=
大家好,今天继续给大家解析这篇文章很有用的代码。
在对单细胞的数据进行可视化的时候,气泡图是很常用的研究工具,但是一般的Seurat的代码给出的气泡2是这样的。
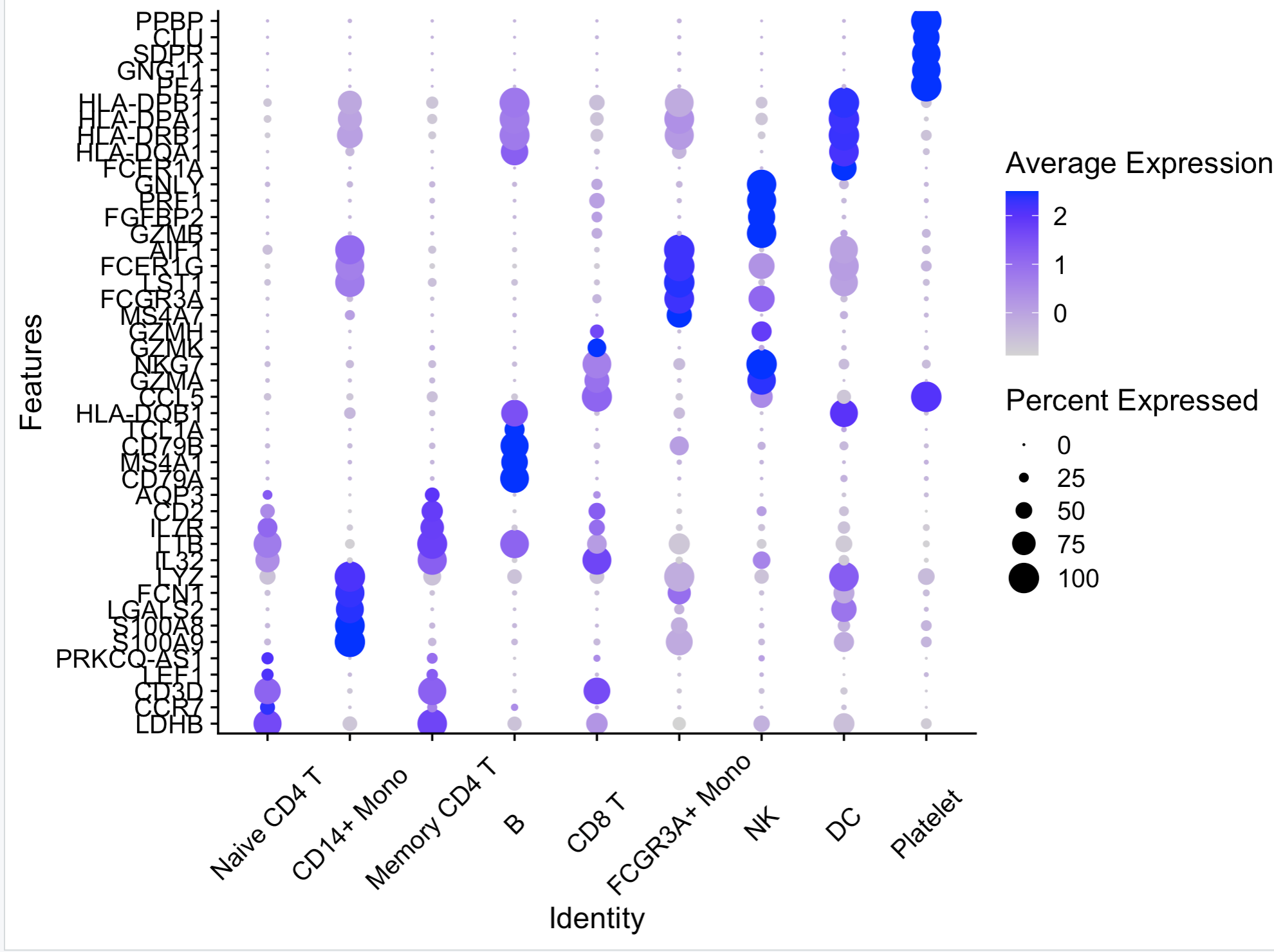
可以发现,无论是配色还是排版都跟CNS的文章相差很远,因此很多人也对这个函数进行了改写,加入了配色等。
因此这篇文章的作者也对代码进行了改写,放入了更多的信息,同时颜色布局也更好看一些。
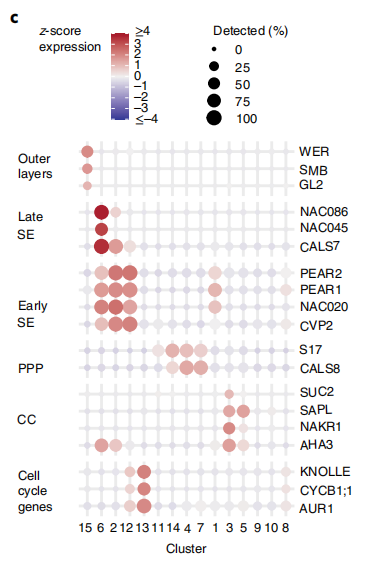
代码解析
// 输入代码内容
# R包的加载
library(SingleCellExperiment)
library(tidyverse)
library(ggpointdensity)
library(patchwork)
# set ggplot2 theme
theme_set(theme_classic() + theme(text = element_text(size = 14)))
# source util functions
source("analysis/functions/utils.R")
# Read Data ---------------------------------------------------------------
# hard filtering
ring <- readRDS("data/processed/SingleCellExperiment/ring_batches_strictfilt.rds")
# order clusters more logically
# 对cluster的顺序进行更改
ring$cluster_mnn_logvst <- factor(ring$cluster_mnn_logvst,
levels = c("8", "10", "9",
"5", "3", "1",
"7", "4", "14", "11",
"13",
"12", "2", "6",
"15"))
# cluster-by-cluster test
# 读取marker gene的表格
cluster_test <- read.csv("data/processed/gene_sets/ring_strictfilt_cluster_markers.csv",
stringsAsFactors = FALSE) %>%
mutate(cluster = factor(cluster))
# curated list of genes
# 对marker gene的名字进行替换
# =前面的是亚群命名,后面是基因名
curated_genes <- list(`Outer Layers` = tribble(~id, ~name,
"AT1G79580", "SMB",
"AT1G79840", "GL2",
"AT5G14750", "WER"),
`SE (early)` = tribble(~id, ~name,
"AT1G05470", "CVP2",
"AT1G54330", "NAC020",
"AT2G37590", "PEAR1",
"AT5G02460", "PEAR2"),
`SE (late)` = tribble(~id, ~name,
"AT1G06490", "CALS7",
"AT5G17260", "NAC086"),
`PPP` = tribble(~id, ~name,
"AT2G22850", "S17",
"AT3G14570", "CALS8"),
`CC` = tribble(~id, ~name,
"AT2G38640", "AT2G38640",
"AT3G12730", "SAPL",
"AT1G22710", "SUC2",
"AT5G02600", "NAKR1",
"AT5G57350", "AHA3"),
`early` = tribble(~id, ~name,
"AT1G29160", "DOF1.5",
"AT2G36400", "GRF3",
"AT5G52870", "MAKR5")) %>%
bind_rows(.id = "tissue")
# Cell type --------------------------------------------------------------
# 对上面的细胞类型进行可视化
# UMAP with clusters
ggplot(getReducedDim(ring, "UMAP30_MNN_logvst"),
aes(V1, V2)) +
geom_point(aes(colour = cluster_mnn_logvst),
show.legend = FALSE) +
geom_point(stat = "centroid", aes(group = cluster_mnn_logvst),
shape = 21, size = 5, fill = "white", alpha = 0.7) +
geom_text(stat = "centroid",
aes(group = cluster_mnn_logvst, label = cluster_mnn_logvst)) +
ggthemes::scale_colour_tableau(palette = "Tableau 20") +
labs(x = "UMAP 1", y = "UMAP 2", colour = "Cluster") +
theme(axis.text = element_blank(), axis.ticks = element_blank()) +
coord_equal() + theme_void()
# plot for paper
p1 <- ring %>%
getReducedDim("UMAP30_MNN_logvst",
genes = curated_genes$id, melted = TRUE,
exprs_values = "logvst", zeros_to_na = FALSE) %>%
left_join(curated_genes, by = "id") %>%
# scale expression for each gene
group_by(id) %>%
mutate(expr_center = (expr - mean(expr))/sd(expr)) %>%
# calculate percentage of cells where the gene is detected per cluster
group_by(cluster_mnn_logvst, id, name, tissue) %>%
mutate(pct_expressing = (sum(expr > 0)/n())*100) %>%
summarise(mean_expr = mean(expr_center),
pct_expressing = unique(pct_expressing)) %>%
ungroup() %>%
# tidy up the tissue column for nicer plotting
# 这一部分我画图的时候没有采用,主要是为了过滤其中的一部分的早期的组织数据
filter(tissue != "early") %>%
mutate(tissue = factor(tissue,
levels = c("CELL\nCYCLE\nGENES",
"CC", "CC\n&\nMSE",
"PPP\n&\nCC", "PPP",
"EARLY\nPSE",
"LATE\nPSE", "OUTER\nLAYERS"))) %>%
mutate(mean_expr = ifelse(abs(mean_expr) > 4, sign(mean_expr)*4, mean_expr)) %>%
# plot
# 这里主要是对ggplot的图进行美化,可以更改limits的大小,来符合自己数据集的内容
ggplot(aes(name, factor(cluster_mnn_logvst))) +
geom_point(aes(colour = mean_expr, size = pct_expressing)) +
facet_grid(~ tissue, scales = "free", space = "free") +
scale_colour_gradient2(low = "#313695", high = "#a50026", mid = "gray94",
limits = c(-4, 4), breaks = seq(-4, 4, by = 1),
labels = c("<= -4", -3, -2, -1, 0, 1, 2, 3, ">= 4")) +
scale_size_continuous(limits = c(0, 100)) +
guides(shape = "none") +
labs(y = "Cluster", x = "", size = "% Detected",
colour = "Z-score\nExpression") +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
pdf("documents/pdf for figures/cell_type_annotation.pdf",
width = 4.68*1.5, height = 3*1.5)
p1 + theme_minimal(base_size = 12) +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
dev.off()总结
单细胞的气泡图可以包含很多信息,基因在不同部位的表达,因此也是单细胞组学文章中常用的方法,用作者的代码后,发现配色和布局相对很合理,整体相对更和谐。在运用到自己的数据集的时候,需要对其中一部分内容进行改写,比如表达量的大小值,图片布局的大小等。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录

