跟着NatureEcology&Evolution学作图:R语言GGally包展示多变量两两相关系数
跟着NatureEcology&Evolution学作图:R语言GGally包展示多变量两两相关系数
生信技能树
发布于 2024-11-21 08:26:50
发布于 2024-11-21 08:26:50
论文
The evolution of human altriciality and brain development in comparative context
https://www.nature.com/articles/s41559-023-02253-z
数据代码
https://figshare.com/articles/dataset/The_evolution_of_human_altriciality_and_brain_development_in_comparative_context/22242724
论文中的数据和代码都有,看论文,然后对应数据和代码,应该能更好的理解论文的内容。今天的推文我们复现一下论文中的Figure3ab
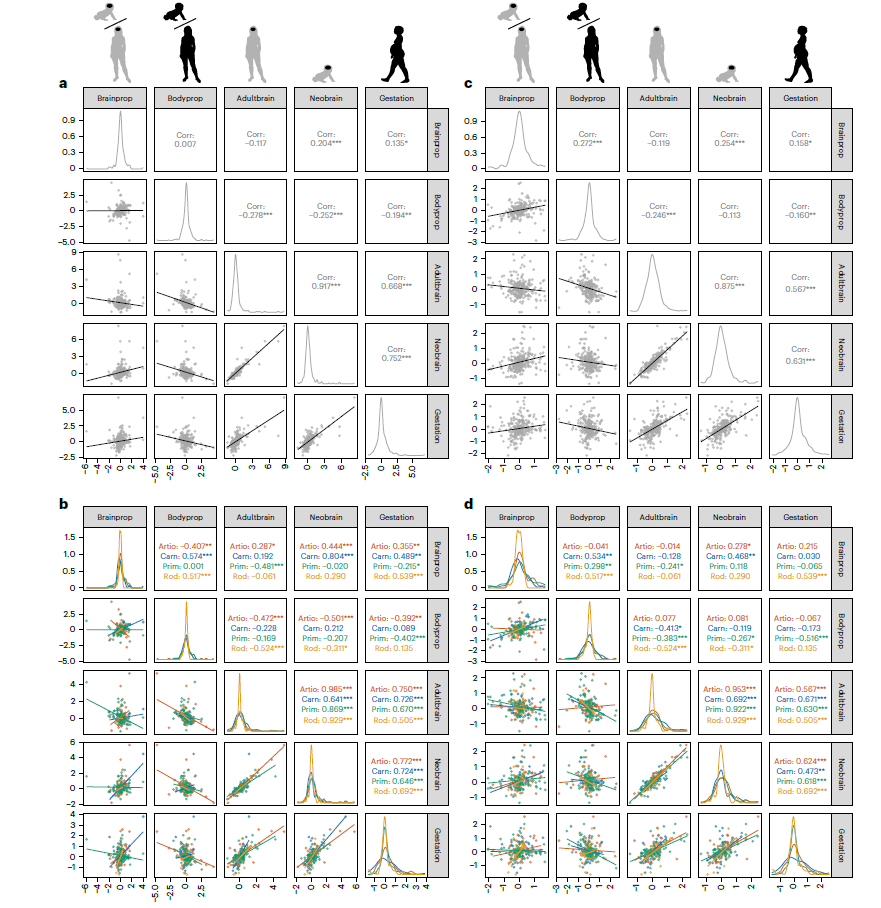
没有获得论文中原始的作图数据,这里我用3个不同品种小麦的一些表型数据做练习,主要学习论文中提供的作图代码
之前的推文也介绍过这个R包,之前发过的推文有
R语言ggplot2画图展示多变量两两之间相关系数~文末留言送书
杂记:ggpairs更改配色;ggplot2极坐标添加直线;seqkit计算fasta序列的长度和gc含量
部分作图数据截图
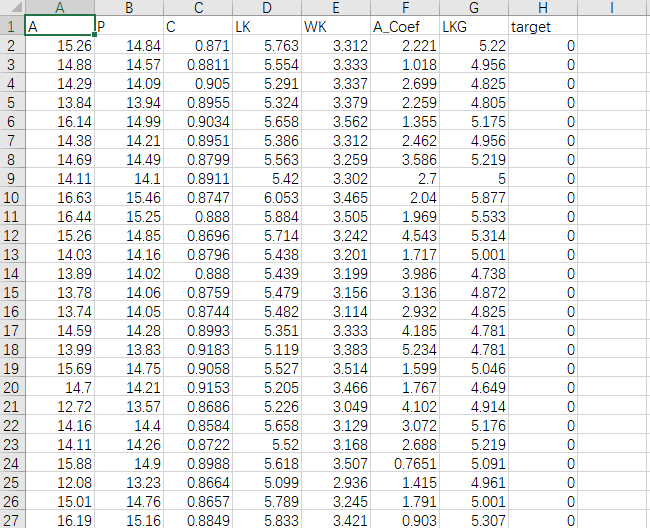
我是用的4.1.0版本的R,没有安装GGally这个包,使用命令install.packages("GGally"),在Rstudio里没有成功,关闭Rstudio,启动R,运行命令可以安装成功。这个是为啥暂时没有想明白
加载需要用到的R包
library(GGally)
library(tidyverse)
library(smplot2)
smplot2这个R包里有一些预设的ggplot2作图可以直接用的主题
读取数据
dat<-read_csv("Seed_Data.csv")
dat %>%
mutate(target=paste("cultivar",target,sep="")) -> dat
dat
这个图主要分为三个部分 右上角upper 对角线diag 和左下角lower
每个好像都可以自定义函数去展示想展示的内容和调节细节
比如论文中的figure3a 定义了左下角的点图和拟合线的颜色
lowerFn <- function(data, mapping, ...) {
p <- ggplot(data = dat, mapping = mapping) +
geom_point(colour = "darkgray",size=0.2,alpha=0.7) +
geom_smooth(method = "lm", color = "black", linewidth=0.3, se=FALSE)
p
}
这里还有很多参数可以写
作图代码
lowerFn <- function(data, mapping, ...) {
p <- ggplot(data = dat, mapping = mapping) +
geom_point(colour = "darkgray",size=0.2,alpha=0.7) +
geom_smooth(method = "lm", color = "black", linewidth=0.3, se=FALSE)
p
}
plot1<-ggpairs(dat[,1:7],
#columns = c("A","P","C","LK","Wk","A_Coef","LKG"),
upper = list(continuous = wrap("cor",size = 5)),
lower = list(continuous = wrap(lowerFn)),
diag = list(continuous = wrap("densityDiag", fill=NA, linewidth=0.3, color="darkgray"))
)
plot1
调节相关系数文本的大小用的是 upper = list(continuous = wrap("cor",size = 5))这行代码中的size参数
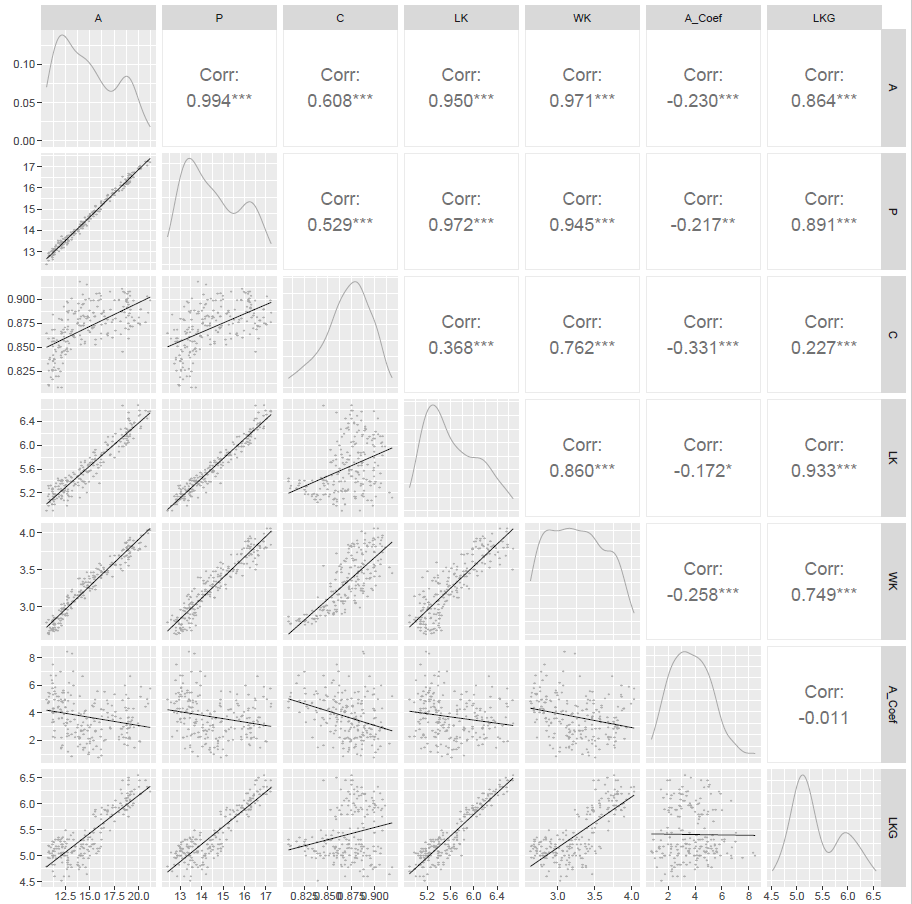
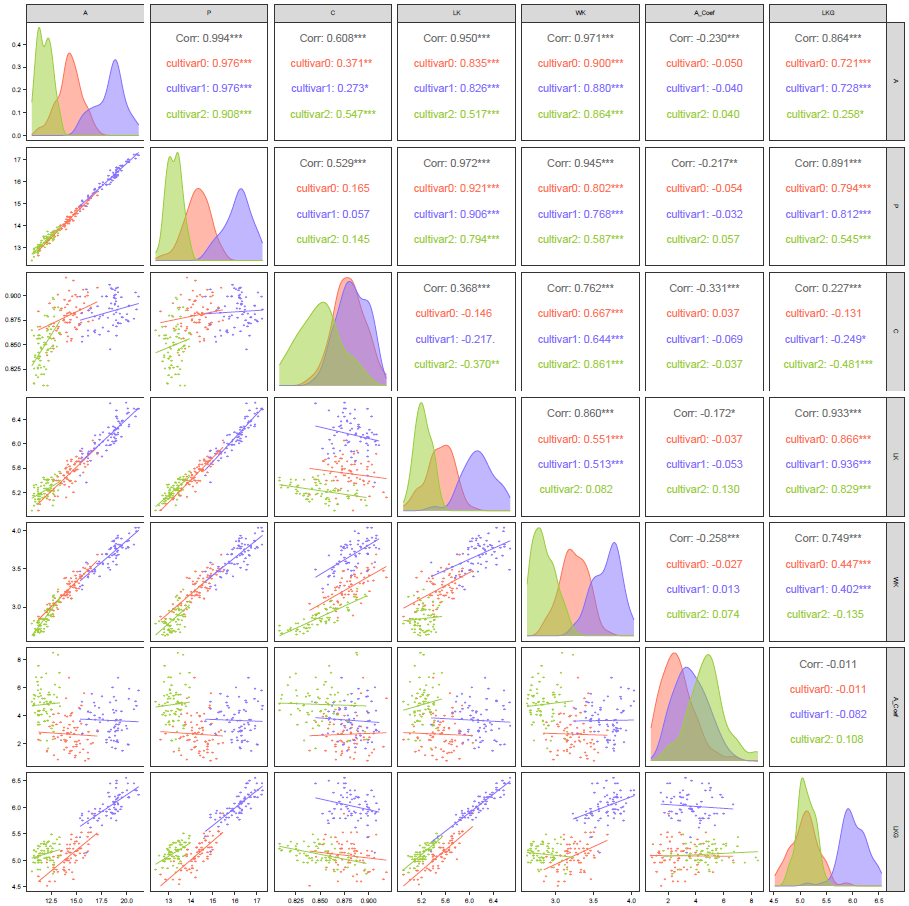
figure3b自定义了对角线和左下角的画图函数
lowerFn2 <- function(data, mapping, ...) {
p <- ggplot(data = dat, mapping = mapping) +
geom_point(size=0.2,alpha=0.7) +
geom_smooth(method = "lm", linewidth=0.3, se = FALSE)
p
}
ggally_mysmooth <- function(data, mapping, ...){
ggplot(data = dat, mapping=mapping) +
geom_density(mapping = aes_string(color="dat$target",fill="dat$target"),
linewidth=0.3,
alpha=0.5)+
theme_bw()
}
作图代码
matrix.corr<-ggpairs(dat[,1:7],
#columns = c("brainprop","bodyprop","adultbrain","neobrain","gestation"),
upper = list(continuous = wrap("cor",size = 5)),
lower = list(continuous = wrap(lowerFn2)),
diag=list(continuous = ggally_mysmooth),
mapping = aes(color = dat$target,fill=dat$target)
)
matrix.corr
matrix.corr +
scale_color_manual(values=c("coral1","lightslateblue","olivedrab3","goldenrod1")) +
scale_fill_manual(values=c("coral1","lightslateblue","olivedrab3","goldenrod1")) +
sm_minimal() +
theme(axis.text=element_text(size = 5)) +
theme(text = element_text(size = 6))
怎么把这两个图组合到一起暂时没搞明白
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2023-12-16,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

