🤩 TrendCatcher | 一把做动态差异表达分析的利剑!~(二)
🤩 TrendCatcher | 一把做动态差异表达分析的利剑!~(二)
生信漫卷
发布于 2024-11-23 09:11:07
发布于 2024-11-23 09:11:07
写在前面
国庆后真的好累啊,感觉这个假期真的意义不大,调休来调休去!~🫠
又转回到创伤了,每个夜班都是无眠啊!~😧
国庆结束后面就没有什么假期了,一直保持工作状态到过年。🙃
㊗️各位今年最后的日子里工作顺利,实验顺利!~🦉
今天继续之前的TrendCatcher,大家做完时序上的差异基因,然后就是想知道这些基因的功能,这就要用到富集分析了。🧐
包内也是提供了可用的函数,今天大家一起看看吧。😘
用到的包
rm(list = ls())
# install.packages("./TrendCatcher_1.0.0.tar.gz", repos = NULL, type = "source")
# devtools::install_github("jaleesr/TrendCatcher", dependencies = TRUE, build_vignettes = FALSE)
library(tidyverse)
library(TrendCatcher)
library(ComplexHeatmap)
示例数据
demo.master.list.path<-system.file("extdata", "BrainMasterList_Symbol.rda", package = "TrendCatcher")
load(demo.master.list.path)
master.list
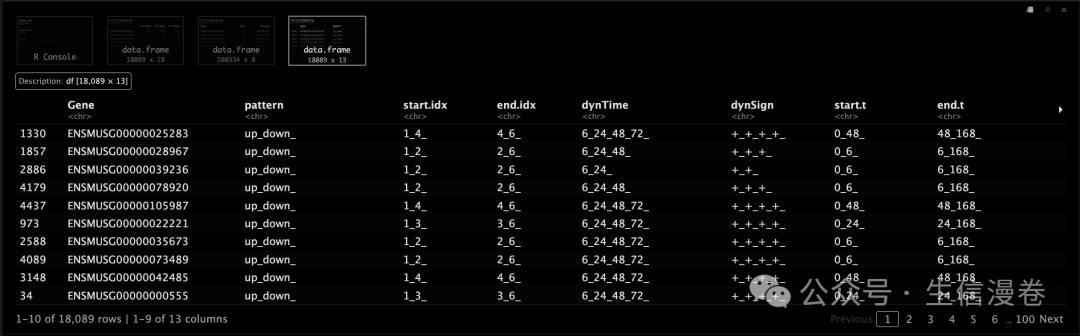
示例结果
这里我们为了节省时间,就把准备好的结果加载进来~!😘
# To save time, directely load from extdata
demo.time.heatmap.path<-system.file("extdata", "Brain_TimeHeatmap.rda", package = "TrendCatcher")
load(demo.time.heatmap.path)
names(time_heatmap)

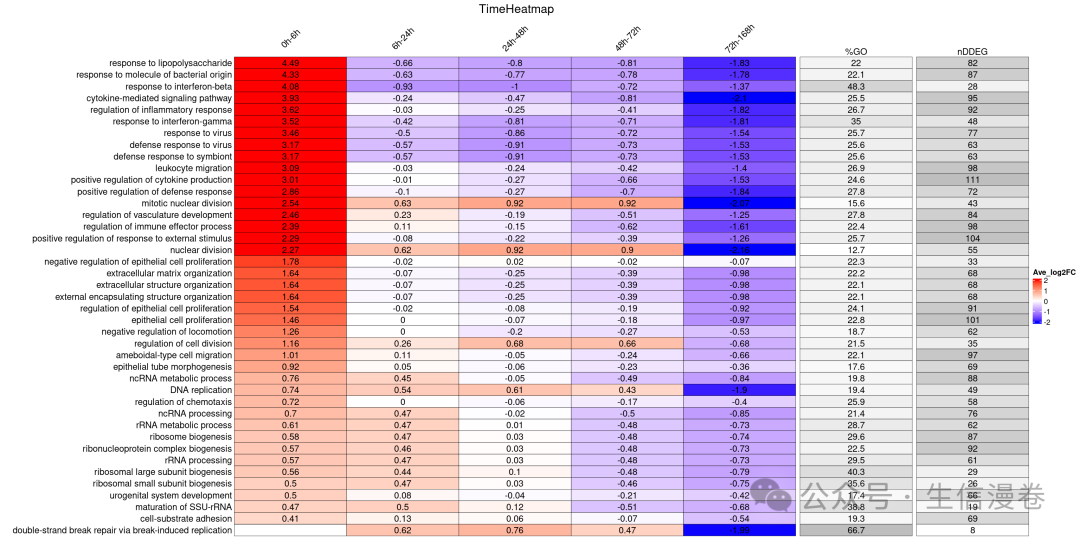
使用 GO 富集分析构建 TimeHeatmap
这里我们使用draw_TimeHeatmap_GO function绘制TimeHeatmap。😘
print(time_heatmap$time.heatmap)
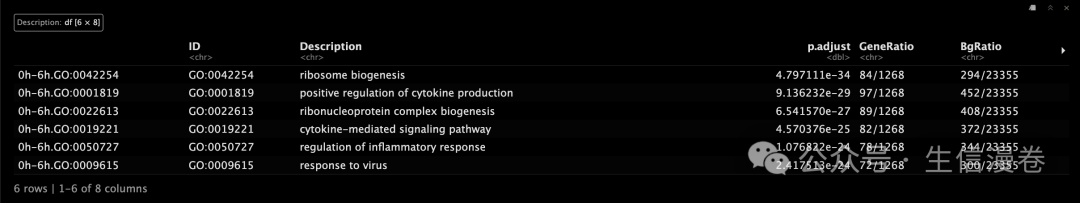
查看富集结果!~
head(time_heatmap$merge.df)
简化GO term并重新绘制TimeHeatmap
有时候你觉得GO term太多了,可以简化一下,然后再绘制TimeHeatmap。😜
go.terms<-unique(time_heatmap$GO.df$Description)[1:5]
time_heatmap_selGO<-draw_TimeHeatmap_selGO(time_heatmap = time_heatmap, sel.go = go.terms, master.list = master.list, GO.perc.thres = 0, nDDEG.thres = 0, save.tiff.path = NA)
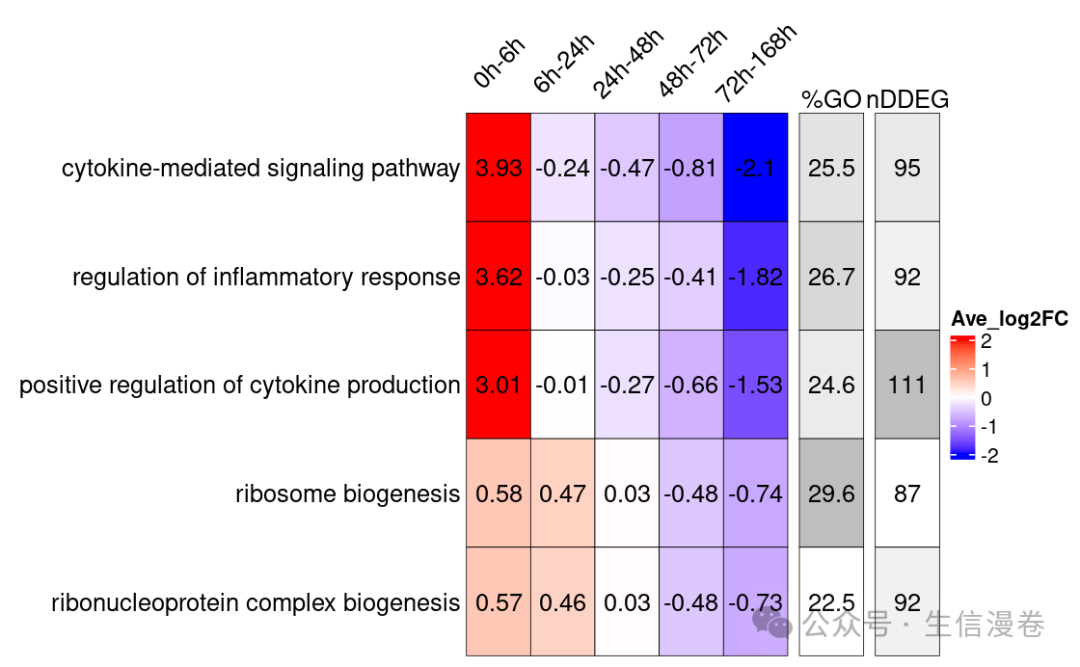
绘制指定GO terms
这里绘制一下我们想要展示的GO terms,并且看一下GO terms下包含的基因。🧬
go.terms<-c("response to lipopolysaccharide",
"response to interferon-beta",
"cytokine-mediated signaling pathway",
"response to interferon-gamma",
"response to virus",
"leukocyte migration",
"mitotic nuclear division",
"regulation of vasculature development",
"extracellular structure organization",
"regulation of epithelial cell proliferation")
gene.GO.df<-draw_GOHeatmap(master.list = master.list, time.window = "0h-6h",
go.terms = go.terms, merge.df = time_heatmap$merge.df,
logFC.thres = 5)
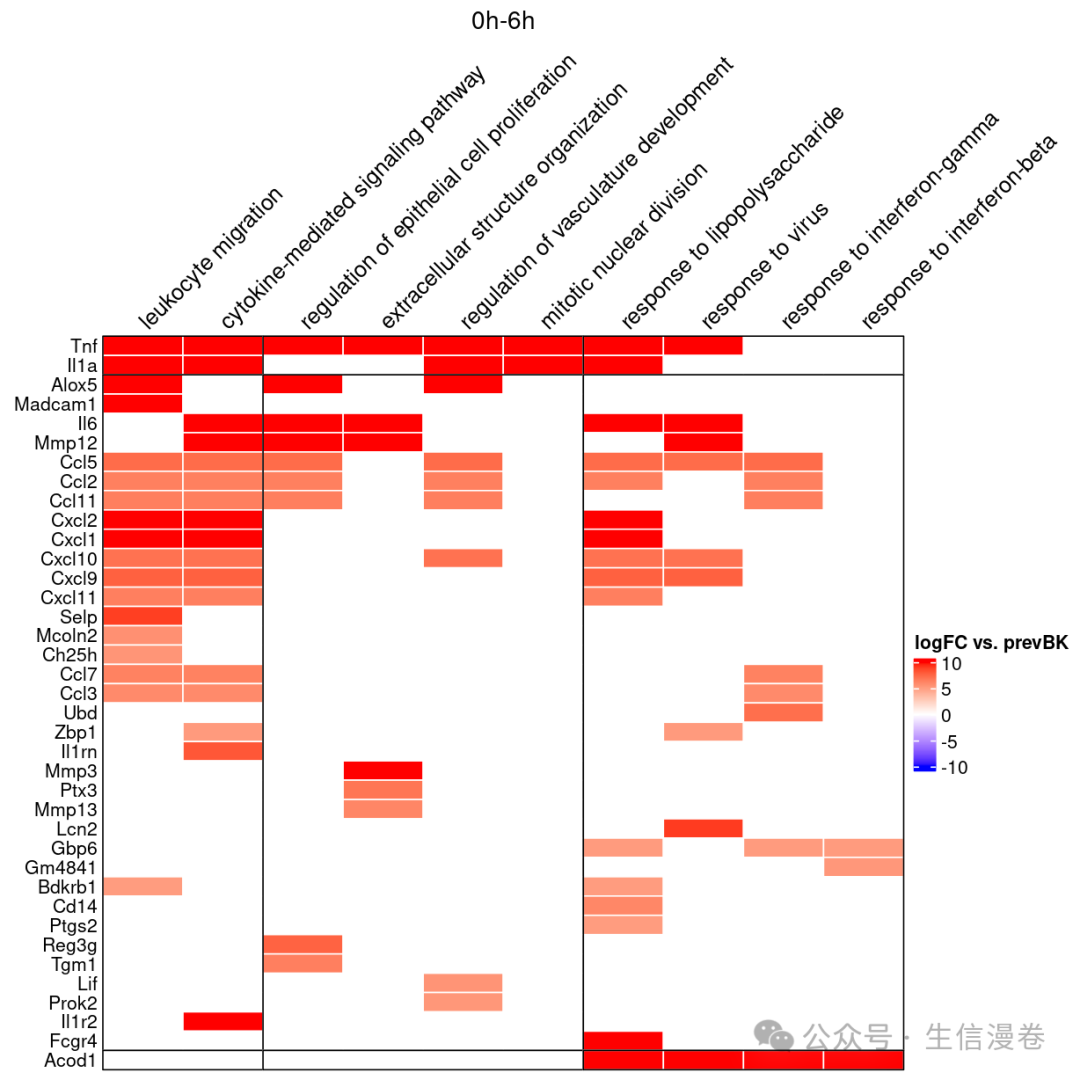
查看详细结果!~😏
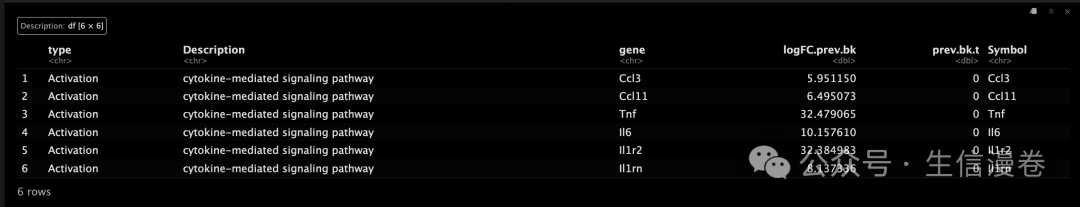
head(gene.GO.df$GOheatmapDat)
最后祝大家早日不卷!~
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2024-10-13,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

