Day9 GEO芯片数据挖掘-以肝癌GSE102079为例
原创Day9 GEO芯片数据挖掘-以肝癌GSE102079为例
原创用户11008504
发布于 2024-05-08 21:21:36
发布于 2024-05-08 21:21:36
GSE102079数据集介绍:
Overall design 整体设计:2006 年至 2011 年间,共有 152 例在东京医科齿科大学医院接受 HCC 根治性肝切除术的患者参加了综合基因表达微阵列分析。从未接受化疗的结直肠癌转移患者中获得的 14 个相邻肝组织作为对照。
1. 下载代码
#打破下载时间的限制,改前60秒,改后10w秒
options(timeout = 100000)
options(scipen = 20)#不要以科学计数法表示
#传统下载方式
library(GEOquery)
eSet = getGEO("GSE102079", destdir = '.', getGPL = F)
eSet = eSet[[1]]
Found 1 file(s)
GSE102079_series_matrix.txt.gz
#(1)提取表达矩阵exp
exp <- exprs(eSet)
dim(exp)
[1] 54613 257
#上步的结果说明表达矩阵有54613行(探针),257列(样本数量),与整体设计样本数量不一致,需注意查看临床信息
range(exp)#看数据范围决定是否需要log,是否有负值,异常值
[1] 2.16531 14.84358
#上步结果表明表达矩阵取值已经取log
boxplot(exp,las = 2) #看是否有异常样本, las = 2横坐标竖直*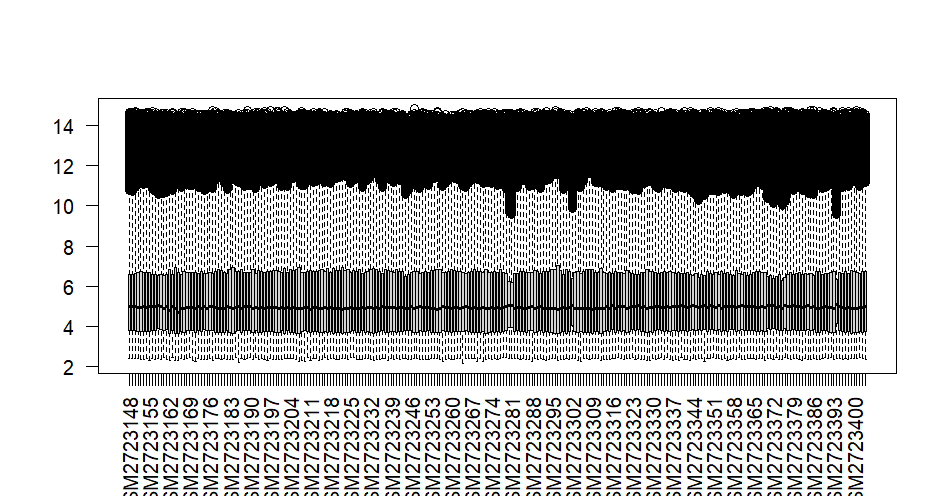
输出结果表明无异常样本及异常值
#(2)提取临床信息
pd <- pData(eSet)
View(pd)#查看临床分组信息,发现有非肿瘤组织不是作者想要分析的
临床信息中样本分组信息
#去除不分析的样本
library(stringr)
k3 = str_detect(pd$source_name_ch1,"non-tumorous");table(k3)
k3
FALSE TRUE
166 91
pd = pd[!k3,]
dim(pd)
[1] 166 34
#上步结果表明符合数据集作者的实验设计14个对照+152个肝癌切除组织
#(3)让exp列名与pd的行名顺序完全一致
p = identical(rownames(pd),colnames(exp));p
[1] FALSE #表明表达矩阵和临床分组信息的样本量不一致,需取交集
if(!p) {
+ s = intersect(rownames(pd),colnames(exp))
+ exp = exp[,s]
+ pd = pd[s,]
+ }
#(4)提取芯片平台编号,后面要根据它来找探针注释
gpl_number <- eSet@annotation;gpl_number
[1] "GPL570"
save(pd,exp,gpl_number,file = "step1output.Rdata")2. Group(实验分组)和ids(探针注释)
rm(list = ls())
load(file = "step1output.Rdata")
# 1.Group----
library(stringr)
k = str_detect(pd$source_name_ch1,"normal");table(k)
k
FALSE TRUE
152 14
Group = ifelse(k,"Normal","Tumor")
# 检查分组信息是否匹配
data.frame(a = pd$source_name_ch1,
b = Group)
# 结果完美匹配 a b
1 Human normal liver #NL001 Normal
2 Human normal liver #NL002 Normal
3 Human normal liver #NL003 Normal
4 Human normal liver #NL004 Normal
5 Human normal liver #NL005 Normal
6 Human normal liver #NL006 Normal
7 Human normal liver #NL007 Normal
8 Human normal liver #NL008 Normal
9 Human normal liver #NL009 Normal
10 Human normal liver #NL010 Normal
11 Human normal liver #NL011 Normal
12 Human normal liver #NL012 Normal
13 Human normal liver #NL013 Normal
14 Human normal liver #NL014 Normal
15 Human liver cancer #LC001, tumorous tissue Tumor
16 Human liver cancer #LC002, tumorous tissue Tumor
17 Human liver cancer #LC003, tumorous tissue Tumor
18 Human liver cancer #LC004, tumorous tissue Tumor
19 Human liver cancer #LC005, tumorous tissue Tumor
20 Human liver cancer #LC006, tumorous tissue Tumor
21 Human liver cancer #LC007, tumorous tissue Tumor
22 Human liver cancer #LC008, tumorous tissue Tumor
23 Human liver cancer #LC009, tumorous tissue Tumor
24 Human liver cancer #LC010, tumorous tissue Tumor
25 Human liver cancer #LC011, tumorous tissue Tumor
26 Human liver cancer #LC012, tumorous tissue Tumor
27 Human liver cancer #LC013, tumorous tissue Tumor
28 Human liver cancer #LC014, tumorous tissue Tumor
29 Human liver cancer #LC015, tumorous tissue Tumor
30 Human liver cancer #LC016, tumorous tissue Tumor
31 Human liver cancer #LC017, tumorous tissue Tumor
32 Human liver cancer #LC018, tumorous tissue Tumor
33 Human liver cancer #LC019, tumorous tissue Tumor
34 Human liver cancer #LC020, tumorous tissue Tumor
35 Human liver cancer #LC021, tumorous tissue Tumor
36 Human liver cancer #LC022, tumorous tissue Tumor
37 Human liver cancer #LC023, tumorous tissue Tumor
38 Human liver cancer #LC024, tumorous tissue Tumor
39 Human liver cancer #LC025, tumorous tissue Tumor
40 Human liver cancer #LC026, tumorous tissue Tumor
41 Human liver cancer #LC027, tumorous tissue Tumor
42 Human liver cancer #LC028, tumorous tissue Tumor
43 Human liver cancer #LC029, tumorous tissue Tumor
44 Human liver cancer #LC030, tumorous tissue Tumor
45 Human liver cancer #LC031, tumorous tissue Tumor
46 Human liver cancer #LC032, tumorous tissue Tumor
47 Human liver cancer #LC033, tumorous tissue Tumor
48 Human liver cancer #LC034, tumorous tissue Tumor
49 Human liver cancer #LC035, tumorous tissue Tumor
50 Human liver cancer #LC036, tumorous tissue Tumor
51 Human liver cancer #LC037, tumorous tissue Tumor
52 Human liver cancer #LC038, tumorous tissue Tumor
53 Human liver cancer #LC039, tumorous tissue Tumor
54 Human liver cancer #LC040, tumorous tissue Tumor
55 Human liver cancer #LC041, tumorous tissue Tumor
56 Human liver cancer #LC042, tumorous tissue Tumor
57 Human liver cancer #LC043, tumorous tissue Tumor
58 Human liver cancer #LC044, tumorous tissue Tumor
59 Human liver cancer #LC045, tumorous tissue Tumor
60 Human liver cancer #LC046, tumorous tissue Tumor
61 Human liver cancer #LC047, tumorous tissue Tumor
62 Human liver cancer #LC048, tumorous tissue Tumor
63 Human liver cancer #LC049, tumorous tissue Tumor
64 Human liver cancer #LC050, tumorous tissue Tumor
65 Human liver cancer #LC051, tumorous tissue Tumor
66 Human liver cancer #LC052, tumorous tissue Tumor
67 Human liver cancer #LC053, tumorous tissue Tumor
68 Human liver cancer #LC054, tumorous tissue Tumor
69 Human liver cancer #LC055, tumorous tissue Tumor
70 Human liver cancer #LC056, tumorous tissue Tumor
71 Human liver cancer #LC057, tumorous tissue Tumor
72 Human liver cancer #LC058, tumorous tissue Tumor
73 Human liver cancer #LC059, tumorous tissue Tumor
74 Human liver cancer #LC060, tumorous tissue Tumor
75 Human liver cancer #LC061, tumorous tissue Tumor
76 Human liver cancer #LC062, tumorous tissue Tumor
77 Human liver cancer #LC063, tumorous tissue Tumor
78 Human liver cancer #LC064, tumorous tissue Tumor
79 Human liver cancer #LC065, tumorous tissue Tumor
80 Human liver cancer #LC066, tumorous tissue Tumor
81 Human liver cancer #LC067, tumorous tissue Tumor
82 Human liver cancer #LC068, tumorous tissue Tumor
83 Human liver cancer #LC069, tumorous tissue Tumor
84 Human liver cancer #LC070, tumorous tissue Tumor
85 Human liver cancer #LC071, tumorous tissue Tumor
86 Human liver cancer #LC072, tumorous tissue Tumor
87 Human liver cancer #LC073, tumorous tissue Tumor
88 Human liver cancer #LC074, tumorous tissue Tumor
89 Human liver cancer #LC075, tumorous tissue Tumor
90 Human liver cancer #LC076, tumorous tissue Tumor
91 Human liver cancer #LC077, tumorous tissue Tumor
92 Human liver cancer #LC078, tumorous tissue Tumor
93 Human liver cancer #LC079, tumorous tissue Tumor
94 Human liver cancer #LC080, tumorous tissue Tumor
95 Human liver cancer #LC081, tumorous tissue Tumor
96 Human liver cancer #LC082, tumorous tissue Tumor
97 Human liver cancer #LC083, tumorous tissue Tumor
98 Human liver cancer #LC084, tumorous tissue Tumor
99 Human liver cancer #LC085, tumorous tissue Tumor
100 Human liver cancer #LC086, tumorous tissue Tumor
101 Human liver cancer #LC087, tumorous tissue Tumor
102 Human liver cancer #LC088, tumorous tissue Tumor
103 Human liver cancer #LC089, tumorous tissue Tumor
104 Human liver cancer #LC090, tumorous tissue Tumor
105 Human liver cancer #LC091, tumorous tissue Tumor
106 Human liver cancer #LC092, tumorous tissue Tumor
107 Human liver cancer #LC093, tumorous tissue Tumor
108 Human liver cancer #LC094, tumorous tissue Tumor
109 Human liver cancer #LC095, tumorous tissue Tumor
110 Human liver cancer #LC096, tumorous tissue Tumor
111 Human liver cancer #LC097, tumorous tissue Tumor
112 Human liver cancer #LC098, tumorous tissue Tumor
113 Human liver cancer #LC099, tumorous tissue Tumor
114 Human liver cancer #LC100, tumorous tissue Tumor
115 Human liver cancer #LC101, tumorous tissue Tumor
116 Human liver cancer #LC102, tumorous tissue Tumor
117 Human liver cancer #LC103, tumorous tissue Tumor
118 Human liver cancer #LC104, tumorous tissue Tumor
119 Human liver cancer #LC105, tumorous tissue Tumor
120 Human liver cancer #LC106, tumorous tissue Tumor
121 Human liver cancer #LC107, tumorous tissue Tumor
122 Human liver cancer #LC108, tumorous tissue Tumor
123 Human liver cancer #LC109, tumorous tissue Tumor
124 Human liver cancer #LC110, tumorous tissue Tumor
125 Human liver cancer #LC111, tumorous tissue Tumor
126 Human liver cancer #LC112, tumorous tissue Tumor
127 Human liver cancer #LC113, tumorous tissue Tumor
128 Human liver cancer #LC114, tumorous tissue Tumor
129 Human liver cancer #LC115, tumorous tissue Tumor
130 Human liver cancer #LC116, tumorous tissue Tumor
131 Human liver cancer #LC117, tumorous tissue Tumor
132 Human liver cancer #LC118, tumorous tissue Tumor
133 Human liver cancer #LC119, tumorous tissue Tumor
134 Human liver cancer #LC120, tumorous tissue Tumor
135 Human liver cancer #LC121, tumorous tissue Tumor
136 Human liver cancer #LC122, tumorous tissue Tumor
137 Human liver cancer #LC123, tumorous tissue Tumor
138 Human liver cancer #LC124, tumorous tissue Tumor
139 Human liver cancer #LC125, tumorous tissue Tumor
140 Human liver cancer #LC126, tumorous tissue Tumor
141 Human liver cancer #LC127, tumorous tissue Tumor
142 Human liver cancer #LC128, tumorous tissue Tumor
143 Human liver cancer #LC129, tumorous tissue Tumor
144 Human liver cancer #LC130, tumorous tissue Tumor
145 Human liver cancer #LC131, tumorous tissue Tumor
146 Human liver cancer #LC132, tumorous tissue Tumor
147 Human liver cancer #LC133, tumorous tissue Tumor
148 Human liver cancer #LC134, tumorous tissue Tumor
149 Human liver cancer #LC135, tumorous tissue Tumor
150 Human liver cancer #LC136, tumorous tissue Tumor
151 Human liver cancer #LC137, tumorous tissue Tumor
152 Human liver cancer #LC138, tumorous tissue Tumor
153 Human liver cancer #LC139, tumorous tissue Tumor
154 Human liver cancer #LC140, tumorous tissue Tumor
155 Human liver cancer #LC141, tumorous tissue Tumor
156 Human liver cancer #LC142, tumorous tissue Tumor
157 Human liver cancer #LC143, tumorous tissue Tumor
158 Human liver cancer #LC144, tumorous tissue Tumor
159 Human liver cancer #LC145, tumorous tissue Tumor
160 Human liver cancer #LC146, tumorous tissue Tumor
161 Human liver cancer #LC147, tumorous tissue Tumor
162 Human liver cancer #LC148, tumorous tissue Tumor
163 Human liver cancer #LC149, tumorous tissue Tumor
164 Human liver cancer #LC150, tumorous tissue Tumor
165 Human liver cancer #LC151, tumorous tissue Tumor
166 Human liver cancer #LC152, tumorous tissue Tumor
#2.探针注释的获取
#捷径
library(tinyarray)
find_anno(gpl_number) #辅助写出找注释的代码,反引号里面的就是注释代码
#`library(hgu133plus2.db);ids <- toTable(hgu133plus2SYMBOL)` and `ids <- AnnoProbe::idmap('GPL570')` are both avaliable
# if you get error by idmap, please try different `type` parameters查AnnoProbe::idmap帮助文档
library(hgu133plus2.db);ids <- toTable(hgu133plus2SYMBOL)
save(exp,Group,ids,file = "step2output.Rdata")3. 主成分分析和基因表达热图
rm(list = ls())
load(file = "step2output.Rdata")
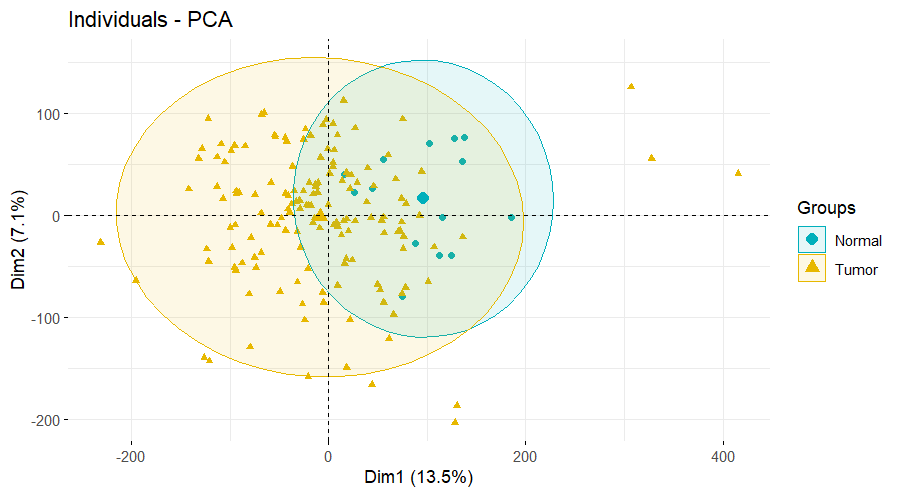
# 1.PCA 图----
dat=as.data.frame(t(exp))
library(FactoMineR)
library(factoextra)
dat.pca <- PCA(dat, graph = FALSE)
fviz_pca_ind(dat.pca,
geom.ind = "point", # show points only (nbut not "text")
col.ind = Group, # color by groups
palette = c("#00AFBB", "#E7B800"),
addEllipses = TRUE, # Concentration ellipses
legend.title = "Groups"
)
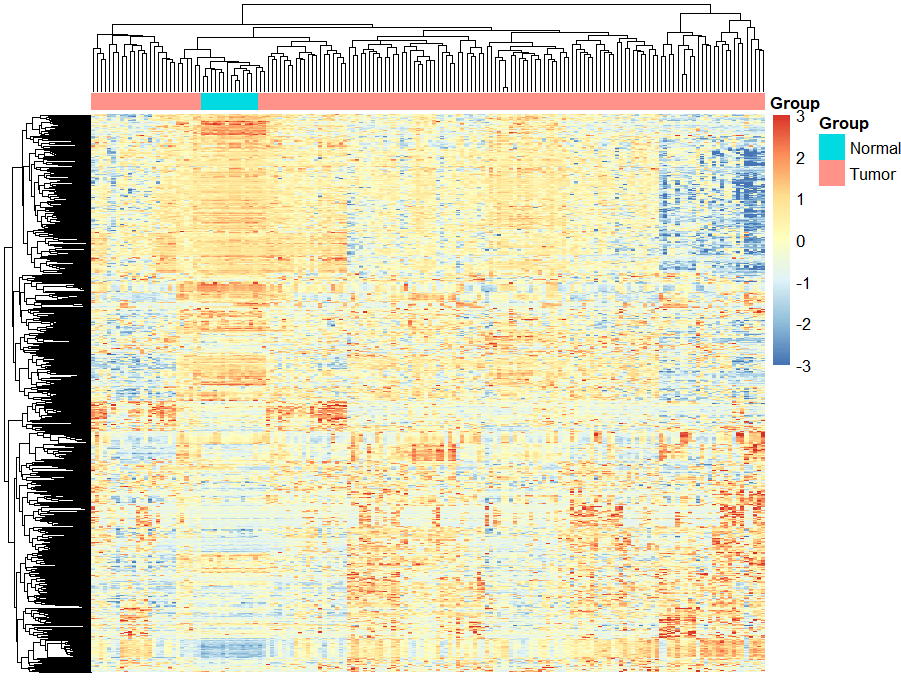
# 2.top 1000 sd 热图----
g = names(tail(sort(apply(exp,1,sd)),1000)) #day7-apply的思考题
n = exp[g,]
library(pheatmap)
annotation_col = data.frame(row.names = colnames(n),
Group = Group)
pheatmap(n,
show_colnames =F,
show_rownames = F,
annotation_col=annotation_col,
scale = "row", #按行标准化,只保留行内差别,不保留行间差别,会把数据范围缩放到大概-5~5之间
breaks = seq(-3,3,length.out = 100) #设置色带分布范围为-3~3之间,超出此范围的数字显示极限颜色
) 
4. 差异分析及火山图绘制
rm(list = ls())
load(file = "step2output.Rdata")
#差异分析
library(limma)
design = model.matrix(~Group)
fit = lmFit(exp,design)
fit = eBayes(fit)
deg = topTable(fit,coef = 2,number = Inf)
#为deg数据框添加几列
#1.加probe_id列,把行名变成一列
library(dplyr)
deg = mutate(deg,probe_id = rownames(deg))
#2.加上探针注释
ids = distinct(ids,symbol,.keep_all = T)
#其他去重方式在zz.去重方式.R
deg = inner_join(deg,ids,by="probe_id")
nrow(deg) #如果行数为0就是你找的探针注释是错的。
#3.加change列,标记上下调基因
logFC_t = 1
p_t = 0.05
#思考,如何使用padj而非p值
k1 = (deg$P.Value < p_t)&(deg$logFC < -logFC_t)
k2 = (deg$P.Value < p_t)&(deg$logFC > logFC_t)
deg = mutate(deg,change = ifelse(k1,"down",ifelse(k2,"up","stable")))
table(deg$change)
#火山图
library(ggplot2)
ggplot(data = deg, aes(x = logFC, y = -log10(P.Value))) +
geom_point(alpha=0.4, size=3.5, aes(color=change)) +
scale_color_manual(values=c("blue", "grey","red"))+
geom_vline(xintercept=c(-logFC_t,logFC_t),lty=4,col="black",linewidth=0.8) +
geom_hline(yintercept = -log10(p_t),lty=4,col="black",linewidth=0.8) +
theme_bw()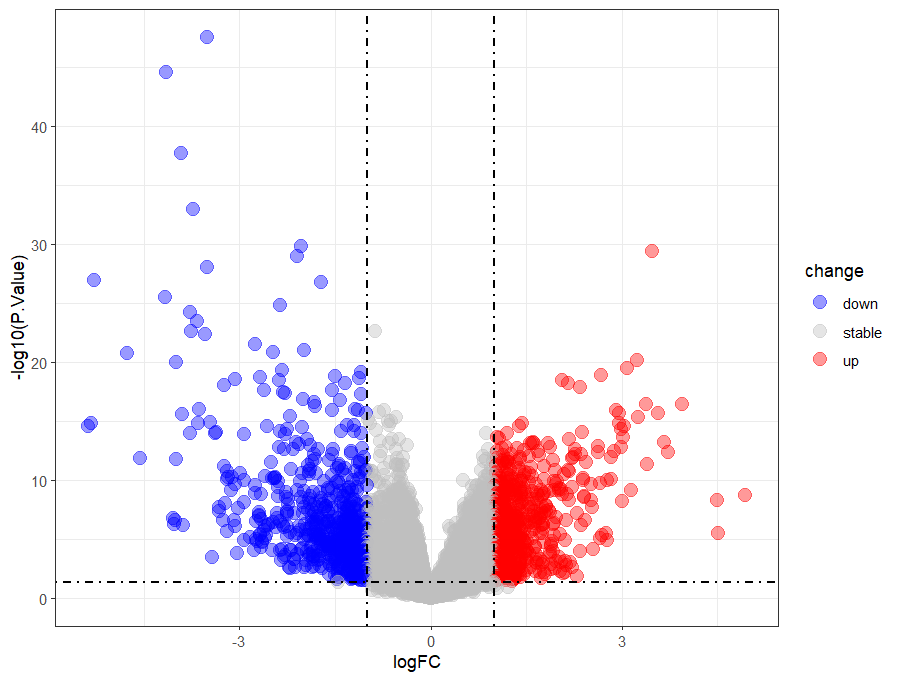
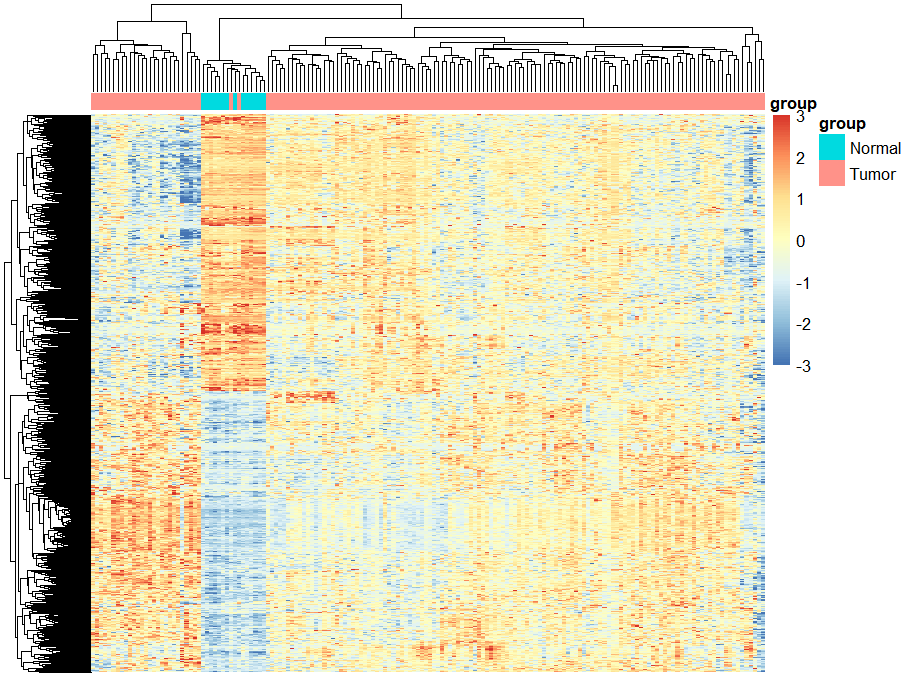
# 差异基因热图----
# 表达矩阵行名替换为基因名
exp = exp[deg$probe_id,]
rownames(exp) = deg$symbol
diff_gene = deg$symbol[deg$change !="stable"]
n = exp[diff_gene,]
library(pheatmap)
annotation_col = data.frame(group = Group)
rownames(annotation_col) = colnames(n)
pheatmap(n,show_colnames =F,
show_rownames = F,
scale = "row",
#cluster_cols = F,
annotation_col=annotation_col,
breaks = seq(-3,3,length.out = 100)
) 
#4.加ENTREZID列,用于富集分析(symbol转entrezid,然后inner_join)
library(clusterProfiler)
library(org.Hs.eg.db)
s2e = bitr(deg$symbol,
fromType = "SYMBOL",
toType = "ENTREZID",
OrgDb = org.Hs.eg.db)#人类,注意物种
#一部分基因没匹配上是正常的。<30%的失败都没事。
#其他物种http://bioconductor.org/packages/release/BiocViews.html#___OrgDb
nrow(deg)
deg = inner_join(deg,s2e,by=c("symbol"="SYMBOL"))
#多了几行少了几行都正常,SYMBOL与ENTREZID不是一对一的。
nrow(deg)
save(exp,Group,deg,logFC_t,p_t,file = "step4output.Rdata")5.GO富集分析
rm(list = ls())
load(file = 'step4output.Rdata')
library(clusterProfiler)
library(ggthemes)
library(org.Hs.eg.db)
library(dplyr)
library(ggplot2)
library(stringr)
library(enrichplot)
#(1)输入数据
gene_diff = deg$ENTREZID[deg$change != "stable"]
#(2)富集
ekk <- enrichKEGG(gene = gene_diff,organism = 'hsa')
ekk <- setReadable(ekk,OrgDb = org.Hs.eg.db,keyType = "ENTREZID")
ego <- enrichGO(gene = gene_diff,OrgDb= org.Hs.eg.db,
ont = "ALL",readable = TRUE)
#setReadable和readable = TRUE都是把富集结果表格里的基因名称转为symbol
class(ekk)
#(3)可视化
barplot(ego, split = "ONTOLOGY") +
facet_grid(ONTOLOGY ~ ., space = "free_y",scales = "free_y")
barplot(ekk)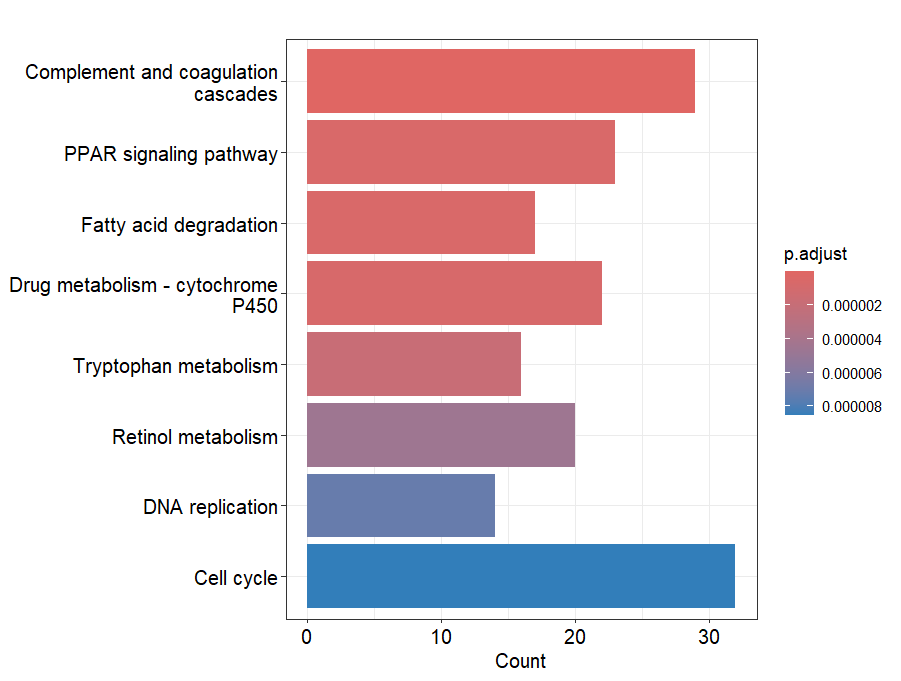
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录

