如何加快编写的python代码:使用空间搜索实现球体接触检测(碰撞)
我正在研究一个空间搜索案例,在这个案例中,我想找到连接的球体。为了达到这个目的,我在每个球体周围搜索中心距离搜索球体中心的距离的球体(maximum sphere直径)。首先,我尝试使用与之相关的方法来实现这一目的,但与等效的numpy方法相比,使用scipy方法要花费更长的时间。对于粒子,我先确定了K-最近的球的数目,然后用cKDTree.query找到它们,这就导致了更多的时间消耗。但是,它比numpy方法慢,即使省略了带有常量值的第一步(在本例中省略第一步是不好的)。--这与我对空间搜索速度的期望是相反的。因此,我尝试使用一些列表循环来加速使用numba prange。Numba运行代码的速度要快一点,但我相信可以通过向量化、使用其他可选的numpy模块或以另一种方式使用numba来优化这段代码以获得更好的性能。为了防止可能的内存泄漏和…,我在所有领域都使用了迭代。球体数目多的地方。
import numpy as np
import numba as nb
from scipy.spatial import cKDTree, distance
# ---------------------------- input data ----------------------------
""" For testing by prepared files:
radii = np.load('a.npy') # shape: (n-spheres, ) must be loaded by np.load('a.npy') or np.loadtxt('radii_large.csv')
poss = np.load('b.npy') # shape: (n-spheres, 3) must be loaded by np.load('b.npy') or np.loadtxt('pos_large.csv', delimiter=',')
"""
rnd = np.random.RandomState(70)
data_volume = 200000
radii = rnd.uniform(0.0005, 0.122, data_volume)
dia_max = 2 * radii.max()
x = rnd.uniform(-1.02, 1.02, (data_volume, 1))
y = rnd.uniform(-3.52, 3.52, (data_volume, 1))
z = rnd.uniform(-1.02, -0.575, (data_volume, 1))
poss = np.hstack((x, y, z))
# --------------------------------------------------------------------
# @nb.jit('float64[:,::1](float64[:,::1], float64[::1])', forceobj=True, parallel=True)
def ends_gap(poss, dia_max):
particle_corsp_overlaps = np.array([], dtype=np.float64)
ends_ind = np.empty([1, 2], dtype=np.int64)
""" using list looping """
# particle_corsp_overlaps = []
# ends_ind = []
# for particle_idx in nb.prange(len(poss)): # by list looping
for particle_idx in range(len(poss)):
unshared_idx = np.delete(np.arange(len(poss)), particle_idx) # <--- relatively high time consumer
poss_without = poss[unshared_idx]
""" # SCIPY method ---------------------------------------------------------------------------------------------
nears_i_ind = cKDTree(poss_without).query_ball_point(poss[particle_idx], r=dia_max) # <--- high time consumer
if len(nears_i_ind) > 0:
dist_i, dist_i_ind = cKDTree(poss_without[nears_i_ind]).query(poss[particle_idx], k=len(nears_i_ind)) # <--- high time consumer
if not isinstance(dist_i, float):
dist_i[dist_i_ind] = dist_i.copy()
""" # NUMPY method --------------------------------------------------------------------------------------------
lx_limit_idx = poss_without[:, 0] <= poss[particle_idx][0] + dia_max
ux_limit_idx = poss_without[:, 0] >= poss[particle_idx][0] - dia_max
ly_limit_idx = poss_without[:, 1] <= poss[particle_idx][1] + dia_max
uy_limit_idx = poss_without[:, 1] >= poss[particle_idx][1] - dia_max
lz_limit_idx = poss_without[:, 2] <= poss[particle_idx][2] + dia_max
uz_limit_idx = poss_without[:, 2] >= poss[particle_idx][2] - dia_max
nears_i_ind = np.where(lx_limit_idx & ux_limit_idx & ly_limit_idx & uy_limit_idx & lz_limit_idx & uz_limit_idx)[0]
if len(nears_i_ind) > 0:
dist_i = distance.cdist(poss_without[nears_i_ind], poss[particle_idx][None, :]).squeeze() # <--- relatively high time consumer
# """ # -------------------------------------------------------------------------------------------------------
contact_check = dist_i - (radii[unshared_idx][nears_i_ind] + radii[particle_idx])
connected = contact_check[contact_check <= 0]
particle_corsp_overlaps = np.concatenate((particle_corsp_overlaps, connected))
""" using list looping """
# if len(connected) > 0:
# for value_ in connected:
# particle_corsp_overlaps.append(value_)
contacts_ind = np.where([contact_check <= 0])[1]
contacts_sec_ind = np.array(nears_i_ind)[contacts_ind]
sphere_olps_ind = np.where((poss[:, None] == poss_without[contacts_sec_ind][None, :]).all(axis=2))[0] # <--- high time consumer
ends_ind_mod_temp = np.array([np.repeat(particle_idx, len(sphere_olps_ind)), sphere_olps_ind], dtype=np.int64).T
if particle_idx > 0:
ends_ind = np.concatenate((ends_ind, ends_ind_mod_temp))
else:
ends_ind[0, 0], ends_ind[0, 1] = ends_ind_mod_temp[0, 0], ends_ind_mod_temp[0, 1]
""" using list looping """
# for contacted_idx in sphere_olps_ind:
# ends_ind.append([particle_idx, contacted_idx])
# ends_ind_org = np.array(ends_ind) # using lists
ends_ind_org = ends_ind
ends_ind, ends_ind_idx = np.unique(np.sort(ends_ind_org), axis=0, return_index=True) # <--- relatively high time consumer
gap = np.array(particle_corsp_overlaps)[ends_ind_idx]
return gap, ends_ind, ends_ind_idx, ends_ind_org在我对23000个球体的一次测试中,使用Colab完成了大约400,200和180秒的循环,500.000个球体需要3.5个小时。对于我的项目来说,这些执行时间根本不令人满意,在一个中等数据卷中,领域的数量可能高达1.000.000。我将在我的主代码中多次调用该代码,并寻找在毫秒(尽可能快的速度)内执行该代码的方法。有可能吗??如果有人能根据需要加快代码的速度,我将不胜感激。
备注:
- 此代码必须在3.7+和GPU上使用python执行。
- 此代码必须适用于数据大小,至少是300.000个域。
- 全是矮胖的,多毛的,还有…等效的模块,而不是我的书面模块,使我的代码更快,将被更新。
谨请就以下问题提出任何建议或解释:
- 在这个问题上,哪种方法可以更快一些?
- 为什么在这种情况下,“参与”并不比其他方法更快?在这个问题上,它可能会有帮助?
- 在迭代器方法和矩阵形式方法之间进行选择对我来说是一个令人困惑的问题。迭代方法使用的内存较少,可以由numba和…使用和调整。但是,我认为,与numpy和…这样的矩阵方法(取决于内存限制)没有用处和可比性。巨大的球体数。在这种情况下,也许我可以省略numpy的迭代,但我强烈地认为,由于巨大的矩阵大小操作和内存泄漏,无法处理迭代。
准备的样本测试数据:
回答 5
Stack Overflow用户
发布于 2022-03-13 01:24:34
在前面回答的基础上,我设计了一个高效的算法,它的内存占用和比以前的快得多(特别是在大型数据集上)。话虽如此,但这个算法远比Python和Numba复杂得多。
以前的算法的关键问题是它们设置了一个dia_max 阈值,这个阈值比实际需要的要大得多。实际上,dia_max被设置为最大可能的redius,以确保不会错过任何重叠。问题是,大数据集包含了非常不同大小的球,其中一些是巨大的。这意味着以前的算法是在许多小球周围获取一个非常大的半径。的结果是成千上万的邻居检查每个球,而只有少数人能真正重叠。
有效解决这个问题的一个解决方案是根据它们的大小将球分成不同的组。其思想是首先基于radii对球进行排序,然后将排序后的球拆分成两个组,然后在每个可能的组之间独立地查询邻居,然后合并数据以便应用前面的算法(还有一些额外的优化)。更确切地说,查询是在小球与大球、小球与其他小球、大球与其他大球、大球与小球之间的查询。
加快速度的另一个关键点是使用请求并行中的不同邻居查询。这个解决方案远非完美,因为BallTree对象需要复制,这是效率低下的,但这是强制性的,因为目前CPython中的并行处理方式(即。(吉尔、泡菜等)。使用支持并行请求的包可以绕过CPython固有的限制,但是现有的包似乎没有提供足够有用的接口来解决这个问题,或者优化得不够,不能真正有用。
最后,可以通过删除几乎所有非常昂贵(隐式)数组分配来对Numba代码进行强优化。使用为小数组优化的就地排序算法,还可以显著缩短执行时间(主要是因为Numba的默认实现执行了几个昂贵的分配,并且没有对小数组进行优化)。此外,最终的np.unique操作可以用一个基本循环完全重写,作为主循环,使用增加的In对球进行迭代(因此已经排序了)。
以下是生成的代码:
import numpy as np
import numba as nb
from sklearn.neighbors import BallTree
from joblib import Parallel, delayed
def flatten_neighbours(arr):
sizes = np.fromiter(map(len, arr), count=len(arr), dtype=np.int64)
values = np.concatenate(arr, dtype=np.int64)
return sizes, values
@delayed
def find_neighbours(searched_pts, ref_pts, max_dist):
balltree = BallTree(ref_pts, leaf_size=16, metric='euclidean')
res = balltree.query_radius(searched_pts, r=max_dist)
return flatten_neighbours(res)
def vstack_neighbours(top_infos, bottom_infos):
top_sizes, top_values = top_infos
bottom_sizes, bottom_values = bottom_infos
return np.concatenate([top_sizes, bottom_sizes]), np.concatenate([top_values, bottom_values])
@nb.njit('(Tuple([int64[::1],int64[::1]]), Tuple([int64[::1],int64[::1]]), int64)')
def hstack_neighbours(left_infos, right_infos, offset):
left_sizes, left_values = left_infos
right_sizes, right_values = right_infos
n = left_sizes.size
out_sizes = np.empty(n, dtype=np.int64)
out_values = np.empty(left_values.size + right_values.size, dtype=np.int64)
left_cur, right_cur, out_cur = 0, 0, 0
right_values += offset
for i in range(n):
left, right = left_sizes[i], right_sizes[i]
full = left + right
out_values[out_cur:out_cur+left] = left_values[left_cur:left_cur+left]
out_values[out_cur+left:out_cur+full] = right_values[right_cur:right_cur+right]
out_sizes[i] = full
left_cur += left
right_cur += right
out_cur += full
return out_sizes, out_values
@nb.njit('(int64[::1], int64[::1], int64[::1], int64[::1])')
def reorder_neighbours(in_sizes, in_values, index, reverse_index):
n = reverse_index.size
out_sizes = np.empty_like(in_sizes)
out_values = np.empty_like(in_values)
in_offsets = np.empty_like(in_sizes)
s, cur = 0, 0
for i in range(n):
in_offsets[i] = s
s += in_sizes[i]
for i in range(n):
in_ind = reverse_index[i]
size = in_sizes[in_ind]
in_offset = in_offsets[in_ind]
out_sizes[i] = size
for j in range(size):
out_values[cur+j] = index[in_values[in_offset+j]]
cur += size
return out_sizes, out_values
@nb.njit
def small_inplace_sort(arr):
if len(arr) < 80:
# Basic insertion sort
i = 1
while i < len(arr):
x = arr[i]
j = i - 1
while j >= 0 and arr[j] > x:
arr[j+1] = arr[j]
j = j - 1
arr[j+1] = x
i += 1
else:
arr.sort()
@nb.jit('(float64[:, ::1], float64[::1], int64[::1], int64[::1])')
def compute(poss, radii, neighbours_sizes, neighbours_values):
n, m = neighbours_sizes.size, np.max(neighbours_sizes)
# Big buffers allocated with the maximum size.
# Thank to virtual memory, it does not take more memory can actually needed.
particle_corsp_overlaps = np.empty(neighbours_values.size, dtype=np.float64)
ends_ind_org = np.empty((neighbours_values.size, 2), dtype=np.float64)
in_offset = 0
out_offset = 0
buff1 = np.empty(m, dtype=np.int64)
buff2 = np.empty(m, dtype=np.float64)
buff3 = np.empty(m, dtype=np.float64)
for particle_idx in range(n):
size = neighbours_sizes[particle_idx]
cur = 0
for i in range(size):
value = neighbours_values[in_offset+i]
if value != particle_idx:
buff1[cur] = value
cur += 1
nears_i_ind = buff1[0:cur]
small_inplace_sort(nears_i_ind) # Note: bottleneck of this function
in_offset += size
if len(nears_i_ind) == 0:
continue
x1, y1, z1 = poss[particle_idx]
cur = 0
for i in range(len(nears_i_ind)):
index = nears_i_ind[i]
x2, y2, z2 = poss[index]
dist = np.sqrt((x2 - x1) ** 2 + (y2 - y1) ** 2 + (z2 - z1) ** 2)
contact_check = dist - (radii[index] + radii[particle_idx])
if contact_check <= 0.0:
buff2[cur] = contact_check
buff3[cur] = index
cur += 1
particle_corsp_overlaps[out_offset:out_offset+cur] = buff2[0:cur]
contacts_sec_ind = buff3[0:cur]
small_inplace_sort(contacts_sec_ind)
sphere_olps_ind = contacts_sec_ind
for i in range(cur):
ends_ind_org[out_offset+i, 0] = particle_idx
ends_ind_org[out_offset+i, 1] = sphere_olps_ind[i]
out_offset += cur
# Truncate the views to their real size
particle_corsp_overlaps = particle_corsp_overlaps[:out_offset]
ends_ind_org = ends_ind_org[:out_offset]
assert len(ends_ind_org) % 2 == 0
size = len(ends_ind_org)//2
ends_ind = np.empty((size,2), dtype=np.int64)
ends_ind_idx = np.empty(size, dtype=np.int64)
gap = np.empty(size, dtype=np.float64)
cur = 0
# Find efficiently duplicates (replace np.unique+np.sort)
for i in range(len(ends_ind_org)):
left, right = ends_ind_org[i]
if left < right:
ends_ind[cur, 0] = left
ends_ind[cur, 1] = right
ends_ind_idx[cur] = i
gap[cur] = particle_corsp_overlaps[i]
cur += 1
return gap, ends_ind, ends_ind_idx, ends_ind_org
def ends_gap(poss, radii):
assert poss.size >= 1
# Sort the balls
index = np.argsort(radii)
reverse_index = np.empty(index.size, np.int64)
reverse_index[index] = np.arange(index.size, dtype=np.int64)
sorted_poss = poss[index]
sorted_radii = radii[index]
# Split them in two groups: the small and the big ones
split_ind = len(radii) * 3 // 4
small_poss, big_poss = np.split(sorted_poss, [split_ind])
small_radii, big_radii = np.split(sorted_radii, [split_ind])
max_small_radii = sorted_radii[max(split_ind, 0)]
max_big_radii = sorted_radii[-1]
# Find the neighbours in parallel
result = Parallel(n_jobs=4, backend='threading')([
find_neighbours(small_poss, small_poss, small_radii+max_small_radii),
find_neighbours(small_poss, big_poss, small_radii+max_big_radii ),
find_neighbours(big_poss, small_poss, big_radii+max_small_radii ),
find_neighbours(big_poss, big_poss, big_radii+max_big_radii )
])
small_small_neighbours = result[0]
small_big_neighbours = result[1]
big_small_neighbours = result[2]
big_big_neighbours = result[3]
# Merge the (segmented) arrays in a big one
neighbours_sizes, neighbours_values = vstack_neighbours(
hstack_neighbours(small_small_neighbours, small_big_neighbours, split_ind),
hstack_neighbours(big_small_neighbours, big_big_neighbours, split_ind)
)
# Reverse the indices.
# Note that the results in `neighbours_values` associated to
# `neighbours_sizes[i]` are subsets of `query_radius([poss[i]], r=dia_max)`
# on a `BallTree(poss)`.
res = reorder_neighbours(neighbours_sizes, neighbours_values, index, reverse_index)
neighbours_sizes, neighbours_values = res
# Finally compute the neighbours with a method similar to the
# previous one, but using a much faster optimized code.
return compute(poss, radii, neighbours_sizes, neighbours_values)
result = ends_gap(poss, radii)这是结果(仍然在相同的i5-9600KF机器上):
Small dataset:
- Reference optimized Numba code: 256 ms
- This highly-optimized Numba code: 82 ms
Big dataset:
- Reference optimized Numba code: 42.7 s (take about 7~8 GiB of RAM)
- This highly-optimized Numba code: 4.2 s (take about 1 GiB of RAM)因此,新算法在小数据集上大约快3.1倍(除了以前的优化之外),在大数据集中的速度是的10倍!这比最初发布的算法快3个数量级。
请注意,80%的时间用于BallTree查询(该查询已经大部分是并行的)。主要的Numba计算功能只需12%的时间,超过75%的时间用于对输入指标进行排序。因此,邻居搜索显然是瓶颈。通过将当前的查询拆分到多个较小的查询中,可以对其进行一些改进,但这将使代码更加复杂,只需进行相对较小的改进(例如。速度快1.5倍)。请注意,更复杂的代码更难维护,而且修改容易出错。因此,我认为迁移到本机语言以克服Python的局限性是提高性能的最佳解决方案。尽管如此,编写更快的本机代码来解决这个问题远非简单(除非您找到好的k-d树、八叉树或球树库)。不过,这肯定比进一步优化这段代码要好。
分析
一次分析表明,至少50%的时间在BallTree中,在没有优化的标量循环中,可以使用像AVX-2 (和循环展开)这样的SIMD指令,速度大约快4倍。此外,还可以看到一些多线程问题(顶部的4个线程是some工作人员,浅绿色部分是空闲时间):
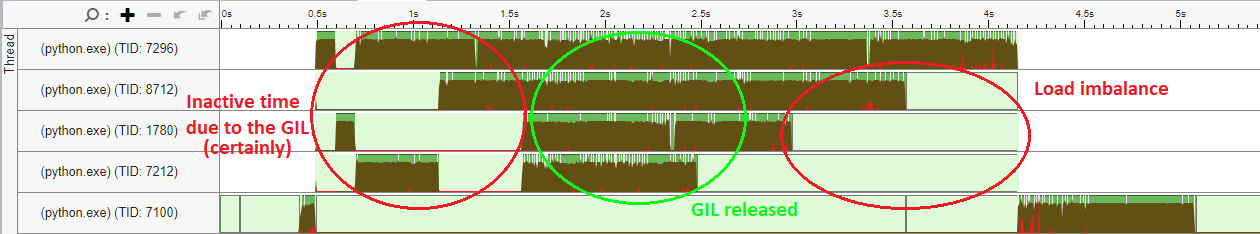
这表明这种实现是次优的。一个可以轻松提高执行时间的方法可能是优化scikit的热点循环-学习BallTree实现。另一种策略可能是更有效地使用线程(可能通过在scikit-learn模块的某些部分释放GIL )。
作为BallTree类的科学学习是用Cython写的 (BallTree是基于DKTree本身基于BinaryTree的)。您可以尝试重新构建计算机上的包,只需调整编译器优化即可。使用参数-O3 -march=native -ffast-math可以使编译器使用更快的SIMD指令和更积极的优化,从而大大加快速度。请注意,使用-ffast-math是不安全的,因为它假设Scikit的代码永远不会使用NaN、Inf或-0值(否则结果是完全未定义的),浮点数操作是关联的(结果不同)。尽管如此,这样一种选择对于改进数字代码的自动矢量化至关重要。
对于GIL,我们可以看到它是在query_radius函数中发布的,但是对于BallTree的构造函数来说,情况似乎并非如此。也许,最简单的解决方案是像西佩那样实现query/query_radius的并行版本。
Stack Overflow用户
发布于 2022-02-16 02:43:47
更新:这个帖子现在被这个新的 (考虑到问题的更新)取代了,它提供了一种基于不同方法的更快的代码。
步骤1:更好的算法
首先,构建k-d树是在O(n log n)时间运行,而查询运行在O(log n)时间,其中n是点数。因此,乍一看,使用k-d树似乎是个好主意。但是,您的代码为每个点构建一个k-d树,从而产生一个O(n² log n)时间。这就是为什么西西的解决方案比其他的慢。问题是,西西没有提供一种更新k-d树的方法。原来是有效更新k-d树似乎是不可能的。。希望在您的情况下,这不是一个问题:您可以只使用所有的点构建一个k-d树,然后丢弃每个查询结果中不希望出现的当前点。
此外,sphere_olps_ind的计算在O(n² m)时间中运行,其中n是总点数,m是平均邻域数(即。从k-d树查询中检索到的最近点)。假设没有重复的点,那么sphere_olps_ind就等于np.sort(contacts_sec_ind)。后者在O(m log m)中运行,这是非常好的。
此外,在循环中使用np.concatenate在Numpy数组中追加值很慢,因为它为每次迭代创建了一个新的更大的数组。使用列表是个好主意,但直接在列表中追加Numpy数组,然后调用,要快得多。
以下是生成的代码:
def ends_gap(poss, dia_max):
particle_corsp_overlaps = []
ends_ind = [np.empty([1, 2], dtype=np.int64)]
kdtree = cKDTree(poss)
for particle_idx in range(len(poss)):
# Find the nearest point including the current one and
# then remove the current point from the output.
# The distances can be computed directly without a new query.
cur_point = poss[particle_idx]
nears_i_ind = np.array(kdtree.query_ball_point(cur_point, r=dia_max), dtype=np.int64)
assert len(nears_i_ind) > 0
if len(nears_i_ind) <= 1:
continue
nears_i_ind = nears_i_ind[nears_i_ind != particle_idx]
dist_i = distance.cdist(poss[nears_i_ind], cur_point[None, :]).squeeze()
contact_check = dist_i - (radii[nears_i_ind] + radii[particle_idx])
connected = contact_check[contact_check <= 0]
particle_corsp_overlaps.append(connected)
contacts_ind = np.where([contact_check <= 0])[1]
contacts_sec_ind = nears_i_ind[contacts_ind]
sphere_olps_ind = np.sort(contacts_sec_ind)
ends_ind_mod_temp = np.array([np.repeat(particle_idx, len(sphere_olps_ind)), sphere_olps_ind], dtype=np.int64).T
if particle_idx > 0:
ends_ind.append(ends_ind_mod_temp)
else:
ends_ind[0][:] = ends_ind_mod_temp[0, 0], ends_ind_mod_temp[0, 1]
ends_ind_org = np.concatenate(ends_ind)
ends_ind, ends_ind_idx = np.unique(np.sort(ends_ind_org), axis=0, return_index=True) # <--- relatively high time consumer
gap = np.concatenate(particle_corsp_overlaps)[ends_ind_idx]
return gap, ends_ind, ends_ind_idx, ends_ind_org步骤2:优化
首先,通过向query_ball_point方法提供poss并指定参数workers=-1,可以在并行中的所有点上同时执行poss调用。但是,请注意,这需要更多的内存。
此外,Numba可以大大加快计算速度。主要改进的部分是计算距离和创建许多不必要的临时数组,以及使用Numpy数组直接索引而不是list的附加(因为输出数组的有界大小可以在query_ball_point调用之后知道)。
下面是使用Numba优化代码的一个简单示例:
@nb.jit('(float64[:, ::1], int64[::1], int64[::1], float64)')
def compute(poss, all_neighbours, all_neighbours_sizes, dia_max):
particle_corsp_overlaps = []
ends_ind_lst = [np.empty((1, 2), dtype=np.int64)]
an_offset = 0
for particle_idx in range(len(poss)):
cur_point = poss[particle_idx]
cur_len = all_neighbours_sizes[particle_idx]
nears_i_ind = all_neighbours[an_offset:an_offset+cur_len]
an_offset += cur_len
assert len(nears_i_ind) > 0
if len(nears_i_ind) <= 1:
continue
nears_i_ind = nears_i_ind[nears_i_ind != particle_idx]
dist_i = np.empty(len(nears_i_ind), dtype=np.float64)
# Compute the distances
x1, y1, z1 = poss[particle_idx]
for i in range(len(nears_i_ind)):
x2, y2, z2 = poss[nears_i_ind[i]]
dist_i[i] = np.sqrt((x2-x1)**2 + (y2-y1)**2 + (z2-z1)**2)
contact_check = dist_i - (radii[nears_i_ind] + radii[particle_idx])
connected = contact_check[contact_check <= 0]
particle_corsp_overlaps.append(connected)
contacts_ind = np.where(contact_check <= 0)
contacts_sec_ind = nears_i_ind[contacts_ind]
sphere_olps_ind = np.sort(contacts_sec_ind)
ends_ind_mod_temp = np.empty((len(sphere_olps_ind), 2), dtype=np.int64)
for i in range(len(sphere_olps_ind)):
ends_ind_mod_temp[i, 0] = particle_idx
ends_ind_mod_temp[i, 1] = sphere_olps_ind[i]
if particle_idx > 0:
ends_ind_lst.append(ends_ind_mod_temp)
else:
tmp = ends_ind_lst[0]
tmp[:] = ends_ind_mod_temp[0, :]
return particle_corsp_overlaps, ends_ind_lst
def ends_gap(poss, dia_max):
kdtree = cKDTree(poss)
tmp = kdtree.query_ball_point(poss, r=dia_max, workers=-1)
all_neighbours = np.concatenate(tmp, dtype=np.int64)
all_neighbours_sizes = np.array([len(e) for e in tmp], dtype=np.int64)
particle_corsp_overlaps, ends_ind_lst = compute(poss, all_neighbours, all_neighbours_sizes, dia_max)
ends_ind_org = np.concatenate(ends_ind_lst)
ends_ind, ends_ind_idx = np.unique(np.sort(ends_ind_org), axis=0, return_index=True)
gap = np.concatenate(particle_corsp_overlaps)[ends_ind_idx]
return gap, ends_ind, ends_ind_idx, ends_ind_org
ends_gap(poss, dia_max)性能分析
以下是我的6核计算机(带有i5-9600KF处理器)在小型数据集上的性能结果:
Initial code with Scipy: 259 s
Initial default code with Numpy: 112 s
Optimized algorithm: 1.37 s
Final optimized code: 0.22 s不幸的是,Scipy树太大了,无法用大数据集在我的机器上存储。
因此,具有高效算法的Numba实现比最初的Numpy实现快~510倍,比初始Scipy实现快1200倍。
Numba代码可以进一步优化,但请注意,Numba compute调用在我的机器上占用的时间不到25%。np.unique调用是最昂贵的,但要使其更快并不容易。很大一部分时间都花在了从数据的转换上,但是只要使用了该代码,这段代码是强制性的。因此,代码可以改进一点(例如。使用高级的Numba优化,但是如果您需要更快的代码,那么您需要使用本地语言,比如C++和高度优化的并行k树实现。我期望一个非常优化的本地代码是一个数量级的更快,但不会更多。我几乎不相信大数据集在我的机器上可以在不到10毫秒的时间内计算出来,不管实现如何。
备注
注意,gap与提供的函数不同(其他值保持不变)。然而,同样的事情发生在初始的Scipy方法和Numpy的方法之间。这似乎来自于诸如nears_i_ind和dist_i这样的变量的排序,这两个变量是由西西未定义的,并以一种非平凡的方式改变gap结果(而不仅仅是gap的顺序)。我不知道这是否初步实施的问题。正因为如此,比较不同实现的正确性要困难得多。
forceobj不应在生产中使用,因为文档指出,这仅用于测试目的。
Stack Overflow用户
发布于 2022-03-07 17:16:27
通过将查询半径固定在最大球半径的两倍,您将创建许多虚假的“冲突”来过滤。
下面的Python通过使用第四维来提高kd树查询的选择性,从而实现了相对于您的答案的显着加速。半径r的每个欧几里德球都被半径为r,√d的L1球过逼近,其中d是维数(3 )。L1球在三维碰撞中的测试变成了在4d内点在固定的L1距离内的测试。
如果切换到较低级别的语言,则可以通过更改kd-tree实现以使用组合L2+L1度量来避免单独的筛选步骤。
import numpy as np
from scipy import spatial
from timeit import default_timer
def load_data():
centers = np.loadtxt("pos_large.csv", delimiter=",")
radii = np.loadtxt("radii_large.csv")
assert radii.shape + (3,) == centers.shape
return centers, radii
def count_contacts(centers, radii):
scaled_centers = centers / np.sqrt(centers.shape[1])
max_radius = radii.max()
tree = spatial.cKDTree(np.c_[scaled_centers, max_radius - radii])
count = 0
for i, x in enumerate(np.c_[scaled_centers, radii - max_radius]):
for j in tree.query_ball_point(x, r=2 * max_radius, p=1):
d = centers[i] - centers[j]
r = radii[i] + radii[j]
if i < j and np.inner(d, d) <= r * r:
count += 1
return count
def main():
centers, radii = load_data()
start = default_timer()
print(count_contacts(centers, radii))
end = default_timer()
print(end - start)
if __name__ == "__main__":
main()https://stackoverflow.com/questions/71104627
复制相似问题

