Pheatmap注释行
Pheatmap注释行
提问于 2022-07-16 10:19:52
因此,我做了一个Deseq分析,然后取下计数文件,应用相同的名称,然后删除安娜值,然后创建一个名为sigs的table/table,然后将其转换为一个数据框架:
sigs <- na.omit(res)
sigs看起来是这样的:
log2 fold change (MLE): condition groupb vs groupa
Wald test p-value: condition groupb vs groupa
DataFrame with 16003 rows and 6 columns
baseMean log2FoldChange lfcSE stat pvalue padj
<numeric> <numeric> <numeric> <numeric> <numeric> <numeric>
ENSSSCG00000048769 82.31674 -0.35837484 0.1217091 -2.9445195 0.00323457 0.0358965
ENSSSCG00000037372 40.49912 0.19133392 0.1472912 1.2990176 0.19393788 0.3612217
ENSSSCG00000027257 1572.05160 0.00319404 0.0743954 0.0429334 0.96575464 0.9791215
ENSSSCG00000029697 494.25472 -0.07424653 0.0665490 -1.1156672 0.26456461 0.4385568
ENSSSCG00000049216 2.54242 -0.42346331 0.5024718 -0.8427604 0.39936246 0.5728141然后我把它转换成一个数据框架:
sigs.df <- as.data.frame(sigs)试图在这里展示:
Description:df [16,003 × 6]
baseMean
<dbl>
log2FoldChange
<dbl>
lfcSE
<dbl>
stat
<dbl>
pvalue
<dbl>
ENSSSCG00000048769 8.231674e+01 -0.3583748397 0.12170911 -2.9445194769 3.234566e-03
ENSSSCG00000037372 4.049912e+01 0.1913339198 0.14729124 1.2990176317 1.939379e-01
ENSSSCG00000027257 1.572052e+03 0.0031940448 0.07439538 0.0429333738 9.657546e-01
ENSSSCG00000029697 4.942547e+02 -0.0742465345 0.06654900 -1.1156672146 2.645646e-01 然后,我尝试将一些参数应用到(Log2fold change和Padj)中。
sigs.df <- sigs.df[(abs(sigs.df$log2FoldChange)>1) & (sigs.df$padj < 0.05),]
sigs.df
Description:df [426 × 6]
baseMean
<dbl>
log2FoldChange
<dbl>
lfcSE
<dbl>
stat
<dbl>
pvalue
<dbl>
padj
<dbl>
18.859565 1.247705 0.4096202 3.046004 2.319046e-03 3.030462e-02
8.702231 -6.199963 1.5519239 -3.995017 6.468949e-05 4.932854e-03
9.466600 -1.535926 0.4899316 -3.134980 1.718657e-03 2.570514e-02
1099.496033 1.547162 0.3705798 4.174976 2.980168e-05 3.222408e-03这里面有426行!然后,我执行规范化、转换,并绘制一个热图:
mat <- counts(dds, normalized = T)[rownames(sigs.df),]
matt(apply(mat,1, scale))dds$condition <- factor(dds$condition, levels = c("Control","Blast"))mat.z <- t(apply(mat,1, scale))
colnames(mat.z) = rownames(coldata)
mat.zlibrary(RColorBrewer)
bluegreen <- c("blue", "green")
pal <- colorRampPalette(bluegreen)(100)par(cex.main=.8)
heatmap(mat.z,cluster_rows = T, cluster_columns = T, column_labels = colnames(mat.z), name = "z-score", col = pal, legend = TRUE,
main = "Heatmap of DEGS Normalized Counts in Pig Samples")
The Output Heattmat is below.
Qu1: It seems to be only displaying a seclection of the genes (Rows labelled on right). How can I get it to display all the genes in detail?
[For thoose wondering, I havent mapped the Ensembl ID’s as there is an issue with Biomart & obtaining the scrofus gene ID’s !]
Qu2: I would like to annotate this with the conditions that each samples (bottom of heatmap) were exposed to. The Sample conditions & runs (Run oone and run 2) are held in the file ‘coldata’ but I am unable to get the heatmap to label/ annotate in this way.
I have seen people call a data frame to do this i./e”
df <- as.data.frame(file$sampleconditions)
then call this with pheatmap (annotation_row = df)..
However I cant seem to get this to work - should I be labelling my sample ID’s with the condition in the same file?
Thanks. Apologies for haphazardness (edited)
:thread:
1
Rob Staruch
5:10 PM
Rplot_Normalised_Counts_Pig_LF2C>1abs, PPadj<0005.png
Rplot_Normalised_Counts_Pig_LF2C>1abs, PPadj<0005.png
:thread:
1
5:10
As an example of the above:
I want to add the annotation row labelling to a pheatmap.
It appears from the tutorial here: https://towardsdatascience.com/pheatmap-draws-pretty-heatmaps-483dab9a3cc
That I can call a data frame in order to do this.
Here is my data frame:
Sample Condition
1 Sample_Run1HR62_S1_Run1 groupa
2 Sample_Run2HR62_S1_Run2 groupa
3 Sample_Run1HR70_S2_Run1 groupa
4 Sample_Run2HR70_S2_Run2 groupa
5 Sample_Run1HR78_S3_Run1 groupa
6 Sample_Run2HR78_S3_Run2 groupa
7 Sample_Run1HR81_S4_Run1 groupa
8 Sample_Run2HR81_S4_Run2 groupa
9 Sample_Run1HR87_S5_Run1 groupa
10 Sample_Run2HR87_S5_Run2 groupa
11 Sample_Run1HR99_S6_Run1 groupa
12 Sample_Run2HR99_S6_Run2 groupa
13 Sample_Run1HR107_S7_Run1 groupa
14 Sample_Run2HR107_S7_Run2 groupa
15 Sample_Run1HR114_S8_Run1 groupa
16 Sample_Run2HR114_S8_Run2 groupa
17 Sample_Run1HR142_S17_Run1 groupa
18 Sample_Run2HR142_S17_Run2 groupa
19 Sample_Run1HR146_S18_Run1 groupa
20 Sample_Run2HR146_S18_Run2 groupa
21 Sample_Run1HR61_S9_Run1 groupb
22 Sample_Run2HR61_S9_Run2 groupb
23 Sample_Run1HR71_S11_Run1 groupb
24 Sample_Run2HR71_S11_Run2 groupb
25 Sample_Run1HR74_S41_Run1 groupb
26 Sample_Run2HR74_S41_Run2 groupb
27 Sample_Run1HR80_S12_Run1 groupb
28 Sample_Run2HR80_S12_Run2 groupb
29 Sample_Run1HR86_S13_Run1 groupb
30 Sample_Run2HR86_S13_Run2 groupb
31 Sample_Run1HR115_S14_Run1 groupb
32 Sample_Run2HR115_S14_Run2 groupb
33 Sample_Run1HR121_S15_Run1 groupb
34 Sample_Run2HR121_S15_Run2 groupb
35 Sample_Run1HR127_S16_Run1 groupb
36 Sample_Run2HR127_S16_Run2 groupb
37 Sample_Run2HR66_S10_Run2 groupb
38 Sample_Run1HR66_S10_Run1 groupb
Here is the r script I am using to generate the Pheatmap:
# Create sample-sample heatmap
sampleDists <- dist(t(assay(rld))) #calculates Euclidean distance. Rld to ensure we have a roughly equal contribution from all genes
sampleDistMatrix <- as.matrix( sampleDists )
rownames(sampleDistMatrix) <- paste( targets$Sample, sep = " - " )
colnames(sampleDistMatrix) <- NULL
colors <- colorRampPalette( rev(brewer.pal(9, "Blues")) )(255)
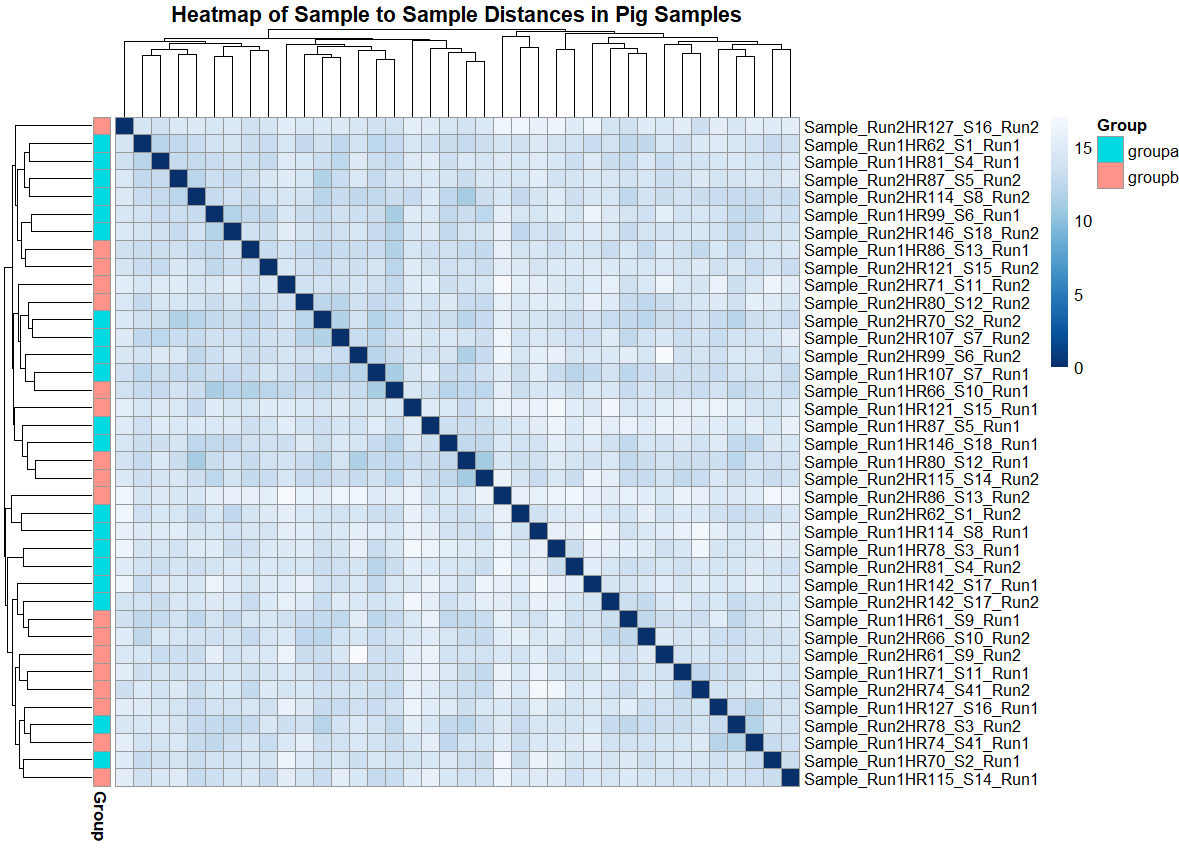
pheatmap(sampleDistMatrix, clustering_distance_rows = sampleDists, clustering_distance_cols = sampleDists,col = colors, main = "Heatmap of Sample to Sample Distances in Pig Samples" )
Here is the same code when I add the ‘annotation_row’ command:
# Create sample-sample heatmap
sampleDists <- dist(t(assay(rld))) #calculates Euclidean distance. Rld to ensure we have a roughly equal contribution from all genes
sampleDistMatrix <- as.matrix( sampleDists )
rownames(sampleDistMatrix) <- paste( targets$Sample, sep = " - " )
colnames(sampleDistMatrix) <- NULL
colors <- colorRampPalette( rev(brewer.pal(9, "Blues")) )(255)
pheatmap(sampleDistMatrix, clustering_distance_rows = sampleDists, clustering_distance_cols = sampleDists,col = colors,annotation_row = targets, main = "Heatmap of Sample to Sample Distances in Pig Samples" )
Here is the error generated from this:
Error in check.length("fill") :
'gpar' element 'fill' must not be length 0
Any help would be greatly appreciatedStack Overflow用户
回答已采纳
发布于 2022-07-16 22:38:19
在我看来,错误是由于targets对象在annotation_row中指定的格式错误造成的。
下面我尝试重现这个错误:
library(pheatmap)
library(RColorBrewer)
targets <- read.table(text="
Sample Group
1 Sample_Run1HR62_S1_Run1 groupa
2 Sample_Run2HR62_S1_Run2 groupa
3 Sample_Run1HR70_S2_Run1 groupa
4 Sample_Run2HR70_S2_Run2 groupa
5 Sample_Run1HR78_S3_Run1 groupa
6 Sample_Run2HR78_S3_Run2 groupa
7 Sample_Run1HR81_S4_Run1 groupa
8 Sample_Run2HR81_S4_Run2 groupa
9 Sample_Run1HR87_S5_Run1 groupa
10 Sample_Run2HR87_S5_Run2 groupa
11 Sample_Run1HR99_S6_Run1 groupa
12 Sample_Run2HR99_S6_Run2 groupa
13 Sample_Run1HR107_S7_Run1 groupa
14 Sample_Run2HR107_S7_Run2 groupa
15 Sample_Run1HR114_S8_Run1 groupa
16 Sample_Run2HR114_S8_Run2 groupa
17 Sample_Run1HR142_S17_Run1 groupa
18 Sample_Run2HR142_S17_Run2 groupa
19 Sample_Run1HR146_S18_Run1 groupa
20 Sample_Run2HR146_S18_Run2 groupa
21 Sample_Run1HR61_S9_Run1 groupb
22 Sample_Run2HR61_S9_Run2 groupb
23 Sample_Run1HR71_S11_Run1 groupb
24 Sample_Run2HR71_S11_Run2 groupb
25 Sample_Run1HR74_S41_Run1 groupb
26 Sample_Run2HR74_S41_Run2 groupb
27 Sample_Run1HR80_S12_Run1 groupb
28 Sample_Run2HR80_S12_Run2 groupb
29 Sample_Run1HR86_S13_Run1 groupb
30 Sample_Run2HR86_S13_Run2 groupb
31 Sample_Run1HR115_S14_Run1 groupb
32 Sample_Run2HR115_S14_Run2 groupb
33 Sample_Run1HR121_S15_Run1 groupb
34 Sample_Run2HR121_S15_Run2 groupb
35 Sample_Run1HR127_S16_Run1 groupb
36 Sample_Run2HR127_S16_Run2 groupb
37 Sample_Run2HR66_S10_Run2 groupb
38 Sample_Run1HR66_S10_Run1 groupb
", header=T)
# Generating a matrix for my example
rld <- matrix(rnorm(100*nr), ncol=nrow(targets))
sampleDists <- dist(t(rld))
sampleDistMatrix <- as.matrix(sampleDists)
rownames(sampleDistMatrix) <- paste(targets$Sample)
colnames(sampleDistMatrix) <- NULL
colors <- colorRampPalette(rev(brewer.pal(9, "Blues")))(255)
pheatmap(sampleDistMatrix, clustering_distance_rows = sampleDists,
clustering_distance_cols = sampleDists, col = colors,
annotation_row = targets,
main="Heatmap of Sample to Sample Distances in Pig Samples")以下是错误:
check.length中的错误(“填充”):'gpar‘元素'fill’不能长度为0
为了解决这个问题,需要对targets进行重新格式化。
首先,targets的行名必须与sampleDistMatrix矩阵相同。
此外,targets必须只有Group列。
rownames(targets) <- rownames(sampleDistMatrix)
targets <- targets[, -1, drop=F]
str(target)
# 'data.frame': 38 obs. of 1 variable:
# $ Group: chr "groupa" "groupa" "groupa" "groupa" ...
pheatmap(sampleDistMatrix, clustering_distance_rows = sampleDists,
clustering_distance_cols = sampleDists, col = colors,
annotation_row = targets,
main="Heatmap of Sample to Sample Distances in Pig Samples")
页面原文内容由Stack Overflow提供。腾讯云小微IT领域专用引擎提供翻译支持
原文链接:
https://stackoverflow.com/questions/73003388
复制相关文章
相似问题

