如何使用一般等价映射将ICD-9码转换成ICD-10码?
来自维基百科
国际疾病分类(ICD)是一种用于流行病学、健康管理和临床目的的全球性诊断工具。
简而言之,ICD是一个代码列表,它有助于以明确的方式描述争议。
我想将使用第9版ICD (ICD-9)的医学数据库转换为使用第10版的ICD-10。人们可以在疾病控制和预防中心(CDC) 网站上找到这两个版本之间的映射文件。
第一个困难产生于这样一个事实:一个ICD-9码可以翻译成一个以上的ICD-10码,这就是某种滑膜。
更困难的问题是,一些代码必须被转换成两个或三个ICD-10码的组合(三个是最大的)。
为了描述这种复杂性,CDC的映射文件看起来与代码8190类似。
| `ICD-9` | `ICD-10` | quality |
|---------+-----------+---------+
| "8190" | "S4291XA" | 10111 |
| "8190" | "S5291XA" | 10111 |
| "8190" | "S4292XA" | 10112 |
| "8190" | "S5292XA" | 10112 |
| "8190" | "S4290XA" | 10121 |
| "8190" | "S5290XA" | 10121 |
| "8190" | "S2220XA" | 10122 |
| "8190" | "S2249XA" | 10123 |quality列表示表示以下标志的标志:
- 第一个数字:1 ==不完全匹配
- 第二个数字:1 ==没有匹配
- 第三位数:1 ==代码必须通过代码组合翻译
- 第四位数:情景
- 第五位:选择列表
场景被细分为目标系统中的两个或多个代码选择列表。这些代码必须作为应用映射中的一个单元连接在一起,以满足源系统中组合代码的等效意义。选择列表包含目标系统中表示源系统中代码的一部分含义的一个或多个代码。必须从场景中的每个选择列表中包含代码,以满足源系统中代码的等效含义。
另一个重要的限制是,我只想保留ICD-10码的前四个字符,所以我用这种方式准备了数据:
| `ICD-9` | `ICD-10` | quality | 4-char ICD-10 | scenario | choice list |
|---------+-----------+---------+---------------+----------+-------------|
| "8190" | "S4291XA" | 10111 | "S429" | 1 | 1 |
| "8190" | "S5291XA" | 10111 | "S529" | 1 | 1 |
| "8190" | "S4292XA" | 10112 | "S429" | 1 | 2 |
| "8190" | "S5292XA" | 10112 | "S529" | 1 | 2 |
| "8190" | "S4290XA" | 10121 | "S429" | 2 | 1 |
| "8190" | "S5290XA" | 10121 | "S529" | 2 | 1 |
| "8190" | "S2220XA" | 10122 | "S222" | 2 | 2 |
| "8190" | "S2249XA" | 10123 | "S224" | 2 | 3 |我还需要使用tidyverse tibble,因为我希望在输出数据中使用嵌套列表。输出数据应该如下所示:
|--------+-------------+----------------------+-------------------------+---------------------------------|
| ICD-9 | Nb of match | One code matches | Two codes matches | Three codes matches |
|--------+-------------+----------------------+-------------------------+---------------------------------|
| "8190" | 6 | list("S429", "S529") | list(c("S429", "S529")) | list(c("S429", "S222", "S224"), |
| | | | | c("S529", "S222", "S224")) |
|--------+-------------+----------------------+-------------------------+---------------------------------| 我们获得了两个单码匹配和一个两个代码匹配,因为当您探索使用截断代码的scenario == 1的场景时,您将获得4种组合选择的方法:
S429 S429 => S429
S429 S529
S529 S429 (identical as the one above)
S529 S529 => S529下面是一个输入数据集:
library(tibble)
input <- structure(list(`ICD-9` = c("00320", "00589", "00589", "01480","01480", "8190", "8190", "8190", "8190", "8190", "8190", "8190", "8190", "36570"), `ICD-10` = c("A0220", "A054", "A058", "A1832", "A1839", "S4291XA", "S5291XA", "S4292XA", "S5292XA", "S4290XA", "S5290XA", "S2220XA", "S2249XA", "NoDx"), quality = c("00000", "10000", "10000", "10000", "10000", "10111", "10111", "10112", "10112", "10121", "10121", "10122", "10123", "11000")), class = c("spec_tbl_df", "tbl_df", "tbl", "data.frame"), row.names = c(NA, -14L), spec = structure(list(cols = list(X1 = structure(list(), class = c("collector_character", "collector")), X2 = structure(list(), class = c("collector_character", "collector")), X3 = structure(list(), class = c("collector_character", "collector"))), default = structure(list(), class = c("collector_guess", "collector")), skip = 0), class = "col_spec"))看上去像是:
# A tibble: 14 x 3
`ICD-9` `ICD-10` quality
<chr> <chr> <chr>
1 00320 A0220 00000
2 00589 A054 10000
3 00589 A058 10000
4 01480 A1832 10000
5 01480 A1839 10000
6 8190 S4291XA 10111
7 8190 S5291XA 10111
8 8190 S4292XA 10112
9 8190 S5292XA 10112
10 8190 S4290XA 10121
11 8190 S5290XA 10121
12 8190 S2220XA 10122
13 8190 S2249XA 10123
14 36570 NoDx 11000 以及预期产出:
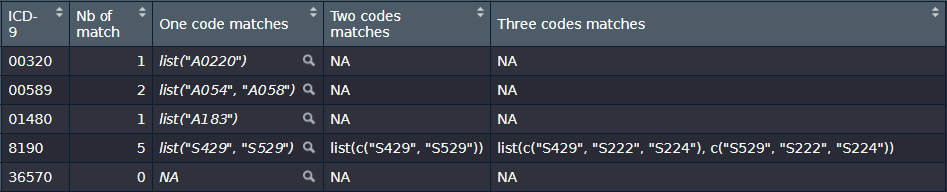
output <- tibble(`ICD-9` = c("00320", "00589", "01480", "8190", "36570"), `Nb of match` = c(1, 2, 1, 5, 0), `One code matches` = list(list("A0220"), list("A054", "A058"), list("A183"), list("S429", "S529"), NA), `Two codes matches` = list(NA, NA, NA, list(c("S429", "S529")), NA), `Three codes matches` = list(NA, NA, NA, list(c("S429", "S222", "S224"), c("S529", "S222", "S224")), NA))看起来是这样的:
回答 1
Stack Overflow用户
发布于 2021-11-21 00:56:12
我在这里的方法是按ICD-9和场景分组,以获得最初的2和3代码匹配。然后压缩这些代码,这样每个ICD-9代码就有一行.然后根据这一重复说明所有可能的重复和从一栏到另一栏的移动。然后把匹配的数量加起来。
我写了许多简短的自定义函数来解释NA和字符(0)等。这给出了您所要求的结果,但这是一个非常小的数据样本,说明这个问题有多复杂。我试着让它一般地工作,但它可能不能。
library(dplyr)
working <- input %>%
mutate(`4-char ICD-10` = substr(`ICD-10`, 1, 4),
scenario = substr(quality, 4, 4),
`choice list` = substr(quality, 5, 5))
expand_across <- function(...) {
do.call(tidyr::expand_grid, list(..., .name_repair = "universal")) %>%
suppressMessages() %>%
setNames(NULL) %>%
as.matrix() %>%
apply(1, list) %>%
unlist(recursive = FALSE)
}
first_not_na <- function(x) {
for (i in seq_along(x)){
if (!is.na(x[i])) return(x[i])
}
return(x[1])
}
fix_groups <- function(x) {
lapply(x, \(y) {
ans <- unique(lapply(y, \(z) sort(unique(z), na.last = TRUE)))
if (identical(ans, list(NA_character_))) ans <- unlist(ans)
ans
})
}
pick_len <- function(x, len) {
ans <- x[sapply(x, length) == len]
ans[identical(ans, character(0))] <- NA_character_
ans
}
list_len <- function(x) {
if_else(!is.list(x), 0L, length(x))
}
by_scen <- working %>%
group_by(`ICD-9`, scenario) %>%
summarize(`Nb of match` = 0L,
`One code matches` = recode(first(substr(quality, 1, 3)),
`000` = list(as.list(`4-char ICD-10`)),
`100` = list(as.list(unique(`4-char ICD-10`))),
.default = list(NA_character_)),
`Two code matches` = recode(first(substr(quality, 3, 3)),
`1` = if (last(`choice list`) == "2")
list(expand_across(
`4-char ICD-10`[`choice list` == "1"],
`4-char ICD-10`[`choice list` == "2"])) else
list(NA_character_),
.default = list(NA_character_)),
`Three code matches` = recode(first(substr(quality, 3, 3)),
`1` = if (last(`choice list`) == "3")
list(expand_across(`4-char ICD-10`[`choice list` == "1"],
`4-char ICD-10`[`choice list` == "2"],
`4-char ICD-10`[`choice list` == "3"])) else
list(NA_character_),
.default = list(NA_character_)),
.groups = 'drop')
output <- by_scen %>%
group_by(`ICD-9`) %>%
summarize(across(!scenario, first_not_na), .groups = "drop") %>%
mutate(`Two code matches` = fix_groups(`Two code matches`)) %>%
mutate(`Three code matches` = fix_groups(`Three code matches`)) %>%
mutate(`One code matches` = coalesce(`One code matches`, lapply(`Two code matches`, pick_len, len = 1))) %>%
mutate(`Two code matches` = lapply(`Two code matches`, pick_len, len = 2)) %>%
mutate(`One code matches` = coalesce(`One code matches`, lapply(`Three code matches`, pick_len, len = 1))) %>%
mutate(`Two code matches` = coalesce(`Two code matches`, lapply(`Three code matches`, pick_len, len = 2))) %>%
mutate(`Three code matches` = lapply(`Three code matches`, pick_len, len = 3)) %>%
mutate(`Nb of match` = sapply(`One code matches`, list_len) +
sapply(`Two code matches`, list_len) +
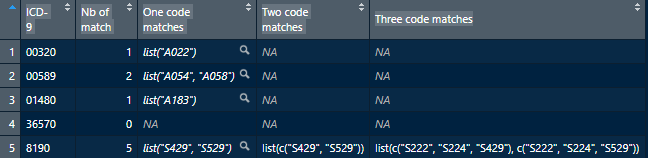
sapply(`Three code matches`, list_len))视图(输出)
https://stackoverflow.com/questions/70038697
复制相似问题

