R中带负值的in图的叠加条形图
R中带负值的in图的叠加条形图
提问于 2021-11-18 21:29:39
这是我的数据集-
df<- data.frame(Comparison=c("Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA",
"SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA",
"Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS",
"HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS"),
LFC_Band=c(-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4,
-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4,
-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4,
-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4),
Number_of_Genes=c(-36,-540,-2198,-3025,-6002,6517,3462,2466, 500,1,-620,-2317,-3318,-2748,-2264,2706,2046,3079,3013,2073,-119,-606,
-1640,-2886,-7451,6944,3147,1592, 267,0,26,0,0, -44,-901 -11699,10969, 754,0,12,0))我试图做的是创建一个堆叠的条形图使用ggplot。
chart_x <- ggplot(data = df, aes(y=(Number_of_Genes),
x=Comparison, group = Comparison))+
geom_col(aes(fill=(as.factor(LFC_Band))))+
scale_fill_brewer(palette = "Spectral")
chart_x结果是-
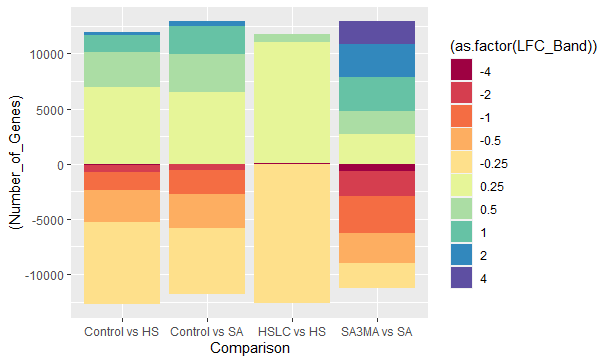
但如果你看到下半部分基本上是翻转的。上半部分是正确的,是4到0.25,而下半部分是从-4开始的。有没有办法把下半身从-0.25开始?总之,我想要的是-
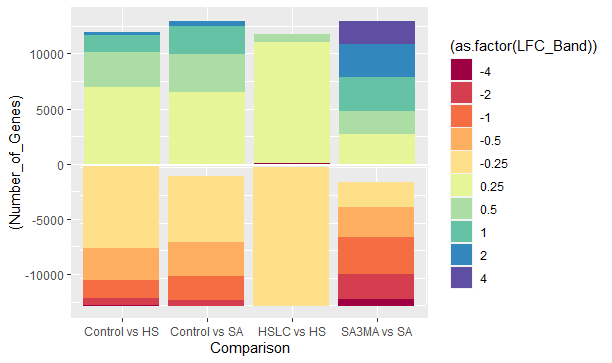
我使用MS油漆来创建它,只是为了展示我正在寻找的东西。
我尝试了其他方法,我把数据分成两部分,但是,它也不起作用。任何帮助都是非常感谢的。:)
回答 2
Stack Overflow用户
回答已采纳
发布于 2021-11-18 22:00:58
要达到你想要的结果,一个选择是:
fill= LFC_Band
- Use values.
- Reverse将
LFC_Band转换为一个因子,然后再将数据传递给ggplot (否则我们丢失了图例中类别的顺序),因此堆栈由
- Drop
group=Comparison分组,其中一个是正的geom_col,一个是负的geom_col,正值
-
drop=FALSE添加到scale_fill_brewer的堆栈顺序为h<215/code>G 216。
另外,我颠倒了填充传说,但是如果你喜欢的话,你可以放弃它。
library(ggplot2)
df$LFC_Band <- factor(df$LFC_Band)
ggplot(data = df, aes(x = Comparison, y = Number_of_Genes, fill = LFC_Band)) +
geom_col(data = df[df$Number_of_Genes > 0, ], position = position_stack(reverse = TRUE)) +
geom_col(data = df[df$Number_of_Genes < 0, ]) +
scale_fill_brewer(palette = "Spectral", drop = FALSE, guide = guide_legend(reverse = TRUE))
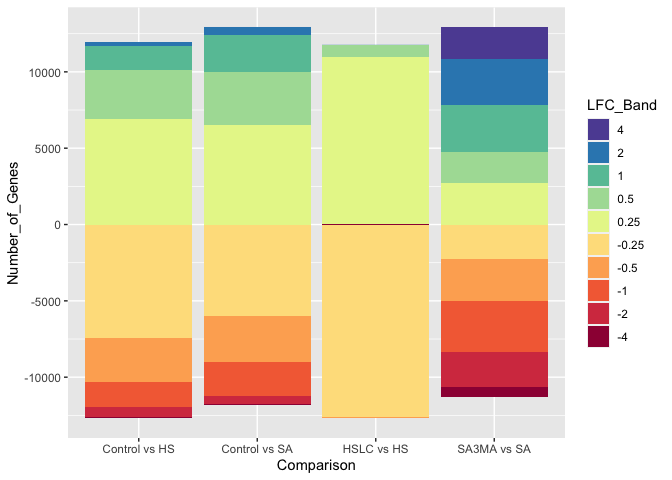
Stack Overflow用户
发布于 2021-11-18 21:58:51
你在找这样的解决方案吗?
library(tidyverse)
df1 <- df %>%
filter(Number_of_Genes>=0)
df2 <- df %>%
filter(Number_of_Genes<0)
ggplot() +
geom_bar(data = df2, aes(x=Comparison, y=Number_of_Genes, group = Comparison, fill=factor(LFC_Band)),stat = "identity") +
geom_bar(data = df1, aes(x=Comparison, y=Number_of_Genes, group = Comparison, fill=factor(LFC_Band)),stat = "identity") +
scale_fill_brewer(palette = "Spectral")

页面原文内容由Stack Overflow提供。腾讯云小微IT领域专用引擎提供翻译支持
原文链接:
https://stackoverflow.com/questions/70026967
复制相关文章
相似问题

