sankeyNetwork不承认定制的colourScale
我正在使用R中的sankeyNetwork创建一个绘图,并且colourScale选项有问题。我创建了一个自定义颜色标度,生成的绘图非常接近于正确,但是在sankeyNetwork函数的某个地方,它破坏了两个组,并将两个组与错误的颜色关联在一起。
这是色标:
colorJS <- "d3.scaleOrdinal() .domain([\" Akatopora spp.\", \"brown fine bryozoan\", \"Bugula neritina\", \"Bugulina californica\", \"Calliarthron tuberculosum\", \"Cauloramphus echinus\", \"Cellaria mandibulata\", \"Celleporaria brunnea\", \"Ceramiales\", \"Cheilostomatida\", \"Chondracanthus spinosus\", \"Chondracanthus spp.\", \"Corallina officinalisvarchilensis\", \"Corallinales\", \"cream encrusting bryozoan\", \"Crisia occidentalis\", \"Cyclostomatida\", \"Diaperoforma californica\", \"Ectoprocta\", \"Gelidiales\", \"Gelidium robustum\", \"Gigartinales\", \"Gloiocladia laciniata\", \"Haywardipora spp.\", \"Jellyella white\", \"Lithothrix aspergillum\", \"Microcladia coulteri\", \"nongeniculate pink\", \"nongeniculate red\", \"other_algae\", \"Parasmittina tubulata\", \"peach encrusting bryozoan\", \"red_fil_alg\", \"red_turf_alg\", \"Rhodophyta\", \"Rhodymenia spp.\", \"Rhodymeniales\", \"Sarcodiotheca furcata\", \"Thalamoporella californica \"]) .range([\" #FF8C00\", \"#FF8C00\", \"#FF8C00\", \"#FF8C00\", \"#DB7093\", \"#FF8C00\", \"#FF8C00\", \"#FF8C00\", \"#DB7093\", \"#FF8C00\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#FF8C00\", \"#FF8C00\", \"#FF8C00\", \"#FF8C00\", \"#FF8C00\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#FF8C00\", \"#FF8C00\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#FFB90F\", \"#FF8C00\", \"#FF8C00\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#DB7093\", \"#FF8C00 \"])"请注意,"red_fil_alg“和"red_turf_alg”与粉红色相关联。
下面是sankeyNetwork的代码:
sankeyNetwork(Links = links, Nodes = nodes,
Source = "source", Target = "target",
Value = "value", NodeID = "name",
fontSize = 20, nodeWidth = 5,
colourScale = colorJS,
LinkGroup="group", NodeGroup = NULL)它产生了这个数字:
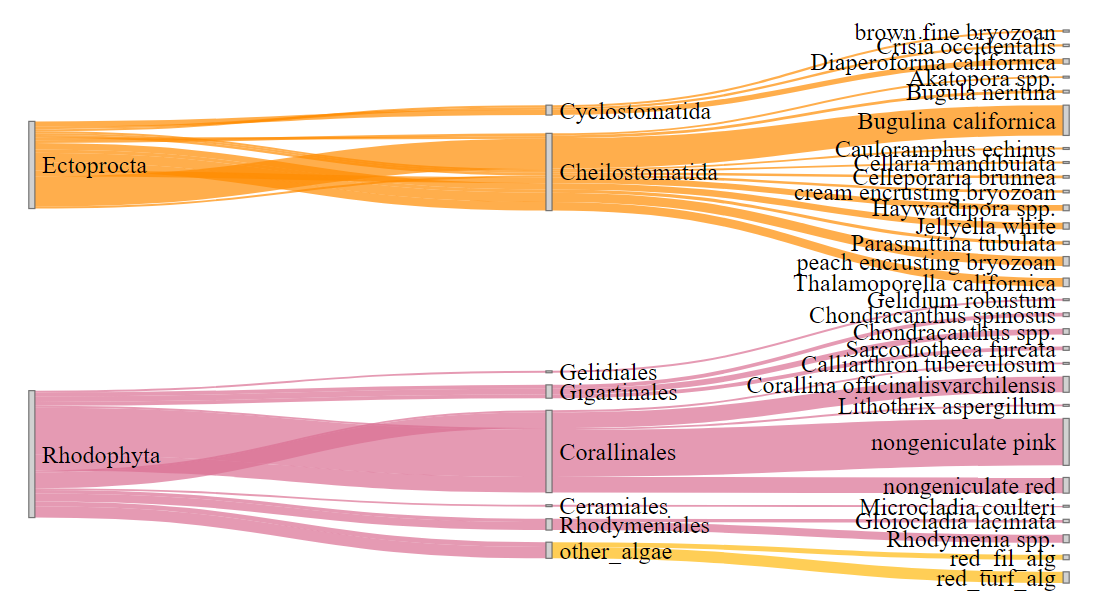
如果不是很明显,"other_algae“、"red_fil_alg”和"red_turf_alg“之间的链接应该是粉红色的,而不是橙色的。我试图通过删除节点名称中的空格来解决这个问题,但这是行不通的。我检查了一下,以确保节点和链接的名称与setdiff()匹配。
以下是链接和节点(如果它们有用的话):
> links
source target value group
1 18 9 1.000000 Ectoprocta
2 18 9 1.500000 Ectoprocta
3 18 9 15.900000 Ectoprocta
4 18 9 1.000000 Ectoprocta
5 18 9 1.000000 Ectoprocta
6 18 9 1.363636 Ectoprocta
7 18 9 1.357143 Ectoprocta
8 18 9 3.025641 Ectoprocta
9 18 9 3.333333 Ectoprocta
10 18 9 1.666667 Ectoprocta
11 18 9 4.934783 Ectoprocta
12 18 9 4.500000 Ectoprocta
13 18 16 1.125000 Ectoprocta
14 18 16 1.250000 Ectoprocta
15 18 16 2.818182 Ectoprocta
16 34 8 1.000000 Rhodophyta
17 34 13 1.000000 Rhodophyta
18 34 13 8.333333 Rhodophyta
19 34 13 1.000000 Rhodophyta
20 34 13 24.655172 Rhodophyta
21 34 13 8.243902 Rhodophyta
22 34 19 1.000000 Rhodophyta
23 34 21 2.000000 Rhodophyta
24 34 21 3.000000 Rhodophyta
25 34 21 2.000000 Rhodophyta
26 34 29 2.555556 Rhodophyta
27 34 29 5.921569 Rhodophyta
28 34 36 1.769231 Rhodophyta
29 34 36 4.187500 Rhodophyta
30 9 0 1.000000 Cheilostomatida
31 9 2 1.500000 Cheilostomatida
32 9 3 15.900000 Cheilostomatida
33 9 5 1.000000 Cheilostomatida
34 9 6 1.000000 Cheilostomatida
35 9 7 1.363636 Cheilostomatida
36 9 14 1.357143 Cheilostomatida
37 9 23 3.025641 Cheilostomatida
38 9 24 3.333333 Cheilostomatida
39 9 30 1.666667 Cheilostomatida
40 9 31 4.934783 Cheilostomatida
41 9 38 4.500000 Cheilostomatida
42 16 1 1.125000 Cyclostomatida
43 16 15 1.250000 Cyclostomatida
44 16 17 2.818182 Cyclostomatida
45 8 26 1.000000 Ceramiales
46 13 4 1.000000 Corallinales
47 13 12 8.333333 Corallinales
48 13 25 1.000000 Corallinales
49 13 27 24.655172 Corallinales
50 13 28 8.243902 Corallinales
51 19 20 1.000000 Gelidiales
52 21 10 2.000000 Gigartinales
53 21 11 3.000000 Gigartinales
54 21 37 2.000000 Gigartinales
55 29 32 2.555556 other_algae
56 29 33 5.921569 other_algae
57 36 22 1.769231 Rhodymeniales
58 36 35 4.187500 Rhodymeniales
> nodes
name
1 Akatopora spp.
2 brown fine bryozoan
3 Bugula neritina
4 Bugulina californica
5 Calliarthron tuberculosum
6 Cauloramphus echinus
7 Cellaria mandibulata
8 Celleporaria brunnea
9 Ceramiales
10 Cheilostomatida
11 Chondracanthus spinosus
12 Chondracanthus spp.
13 Corallina officinalisvarchilensis
14 Corallinales
15 cream encrusting bryozoan
16 Crisia occidentalis
17 Cyclostomatida
18 Diaperoforma californica
19 Ectoprocta
20 Gelidiales
21 Gelidium robustum
22 Gigartinales
23 Gloiocladia laciniata
24 Haywardipora spp.
25 Jellyella white
26 Lithothrix aspergillum
27 Microcladia coulteri
28 nongeniculate pink
29 nongeniculate red
30 other_algae
31 Parasmittina tubulata
32 peach encrusting bryozoan
33 red_fil_alg
34 red_turf_alg
35 Rhodophyta
36 Rhodymenia spp.
37 Rhodymeniales
38 Sarcodiotheca furcata
39 Thalamoporella californica回答 2
Stack Overflow用户
发布于 2021-02-15 12:36:30
我已经测试了对代码的几个更改,以缩小故障范围(见下文)。它们都解决不了这个问题,所以最有可能的原因仍然是库中的一个bug,更具体地说,是关于sankeyNetwork()是如何在有序的域和范围的范围内运行的。我建议您在开发人员的空间上标记此问题。https://github.com/christophergandrud/networkD3/issues?page=2&q=is%3Aissue+is%3Aopen
事实上,有一个类似的问题报道:https://github.com/christophergandrud/networkD3/issues/242解决方案是删除传递给D3域的字符串中的空格。但是,我已经尝试过了,但没有成功:
在"spp"s
- introduced (节点df中基于零的索引)之后,
- 用“-”
- 替换了空格和下划线,从而排除了
的问题。
library(networkD3)
library(tidyverse)
nodes <- data.frame(names = c("Akatopora-spp","brown-fine-bryozoan","Bugula-neritina","Bugulina-californica","Calliarthron-tuberculosum","Cauloramphus-echinus","Cellaria-mandibulata","Celleporaria-brunnea","Ceramiales","Cheilostomatida","Chondracanthus-spinosus","Chondracanthus-spp","Corallina-officinalisvarchilensis","Corallinales","cream-encrusting-bryozoan","Crisia-occidentalis","Cyclostomatida","Diaperoforma-californica","Ectoprocta","Gelidiales","Gelidium-robustum","Gigartinales","Gloiocladia-laciniata","Haywardipora-spp","Jellyella-white","Lithothrix-aspergillum","Microcladia-coulteri","nongeniculate-pink","nongeniculate-red","Other-algae","Parasmittina-tubulata","peach-encrusting-bryozoan","red-fil-algae","red-turf-algae","Rhodophyta","Rhodymenia-spp","Rhodymeniales","Sarcodiotheca-furcata","Thalamoporella-californica"))
links <- data.frame(matrix(c(1,18,9,1.000000,"Ectoprocta",2,18,9,1.500000,"Ectoprocta",3,18,9,15.900000,"Ectoprocta",4,18,9,1.000000,"Ectoprocta",5,18,9,1.000000,"Ectoprocta",6,18,9,1.363636,"Ectoprocta",7,18,9,1.357143,"Ectoprocta",8,18,9,3.025641,"Ectoprocta",9,18,9,3.333333,"Ectoprocta",10,18,9,1.666667,"Ectoprocta",11,18,9,4.934783,"Ectoprocta",12,18,9,4.500000,"Ectoprocta",13,18,16,1.125000,"Ectoprocta",14,18,16,1.250000,"Ectoprocta",15,18,16,2.818182,"Ectoprocta",16,34,8,1.000000,"Rhodophyta",17,34,13,1.000000,"Rhodophyta",18,34,13,8.333333,"Rhodophyta",19,34,13,1.000000,"Rhodophyta",20,34,13,24.655172,"Rhodophyta",21,34,13,8.243902,"Rhodophyta",22,34,19,1.000000,"Rhodophyta",23,34,21,2.000000,"Rhodophyta",24,34,21,3.000000,"Rhodophyta",25,34,21,2.000000,"Rhodophyta",26,34,29,2.555556,"Rhodophyta",27,34,29,5.921569,"Rhodophyta",28,34,36,1.769231,"Rhodophyta",29,34,36,4.187500,"Rhodophyta",30,9,0,1.000000,"Cheilostomatida",31,9,2,1.500000,"Cheilostomatida",32,9,3,15.900000,"Cheilostomatida",33,9,5,1.000000,"Cheilostomatida",34,9,6,1.000000,"Cheilostomatida",35,9,7,1.363636,"Cheilostomatida",36,9,14,1.357143,"Cheilostomatida",37,9,23,3.025641,"Cheilostomatida",38,9,24,3.333333,"Cheilostomatida",39,9,30,1.666667,"Cheilostomatida",40,9,31,4.934783,"Cheilostomatida",41,9,38,4.500000,"Cheilostomatida",42,16,1,1.125000,"Cyclostomatida",43,16,15,1.250000,"Cyclostomatida",44,16,17,2.818182,"Cyclostomatida",45,8,26,1.000000,"Ceramiales",46,13,4,1.000000,"Corallinales",47,13,12,8.333333,"Corallinales",48,13,25,1.000000,"Corallinales",49,13,27,24.655172,"Corallinales",50,13,28,8.243902,"Corallinales",51,19,20,1.000000,"Gelidiales",52,21,10,2.000000,"Gigartinales",53,21,11,3.000000,"Gigartinales",54,21,37,2.000000,"Gigartinales",55,29,32,2.555556,"other_algae",56,29,33,5.921569,"other_algae",57,36,22,1.769231,"Rhodymeniales",58,36,35,4.187500,"Rhodymeniales"), ncol = 5, byrow = TRUE), stringsAsFactors = FALSE) %>%
rename(ID = X1, source = X2, target = X3, value = X4, group = X5) %>%
mutate(value = as.numeric(value)) # input links df
links[links$group == "other-algae",]$group <- "Other-algae"
nodes <- nodes %>%
rowid_to_column(., var = "ID") %>% # introduce index
mutate(ID = ID-1) # zero-based index
colorJS <- "d3.scaleOrdinal() .domain([\"Akatopora-spp\",\"brown-fine-bryozoan\",\"Bugula-neritina\",\"Bugulina-californica\",\"Calliarthron-tuberculosum\",\"Cauloramphus-echinus\",\"Cellaria-mandibulata\",\"Celleporaria-brunnea\",\"Ceramiales\",\"Cheilostomatida\",\"Chondracanthus-spinosus\",\"Chondracanthus-spp\",\"Corallina-officinalisvarchilensis\",\"Corallinales\",\"cream-encrusting-bryozoan\",\"Crisia-occidentalis\",\"Cyclostomatida\",\"Diaperoforma-californica\",\"Ectoprocta\",\"Gelidiales\",\"Gelidium-robustum\",\"Gigartinales\",\"Gloiocladia-laciniata\",\"Haywardipora-spp\",\"Jellyella-white\",\"Lithothrix-aspergillum\",\"Microcladia-coulteri\",\"nongeniculate-pink\",\"nongeniculate-red\",\"Other-algae\",\"Parasmittina-tubulata\",\"peach-encrusting-bryozoan\",\"red-fil-algae\",\"red-turf-algae\",\"Rhodophyta\",\"Rhodymenia-spp\",\"Rhodymeniales\",\"Sarcodiotheca-furcata\",\"Thalamoporella-californica\"]) .range([\" #FF8C00\",\"#FF8C00\",\"#FF8C00\",\"#FF8C00\",\"#DB7093\",\"#FF8C00\",\"#FF8C00\",\"#FF8C00\",\"#DB7093\",\"#FF8C00\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#FF8C00\",\"#FF8C00\",\"#FF8C00\",\"#FF8C00\",\"#FF8C00\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#FF8C00\",\"#FF8C00\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#FFB90F\",\"#FF8C00\",\"#FF8C00\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#DB7093\",\"#FF8C00\"])"
sankeyNetwork(Links = links, Nodes = nodes,
Source = "source", Target = "target",
Value = "value", NodeID = "names",
fontSize = 20, nodeWidth = 5,
colourScale = colorJS,
LinkGroup="group", NodeGroup = NULL)输出:
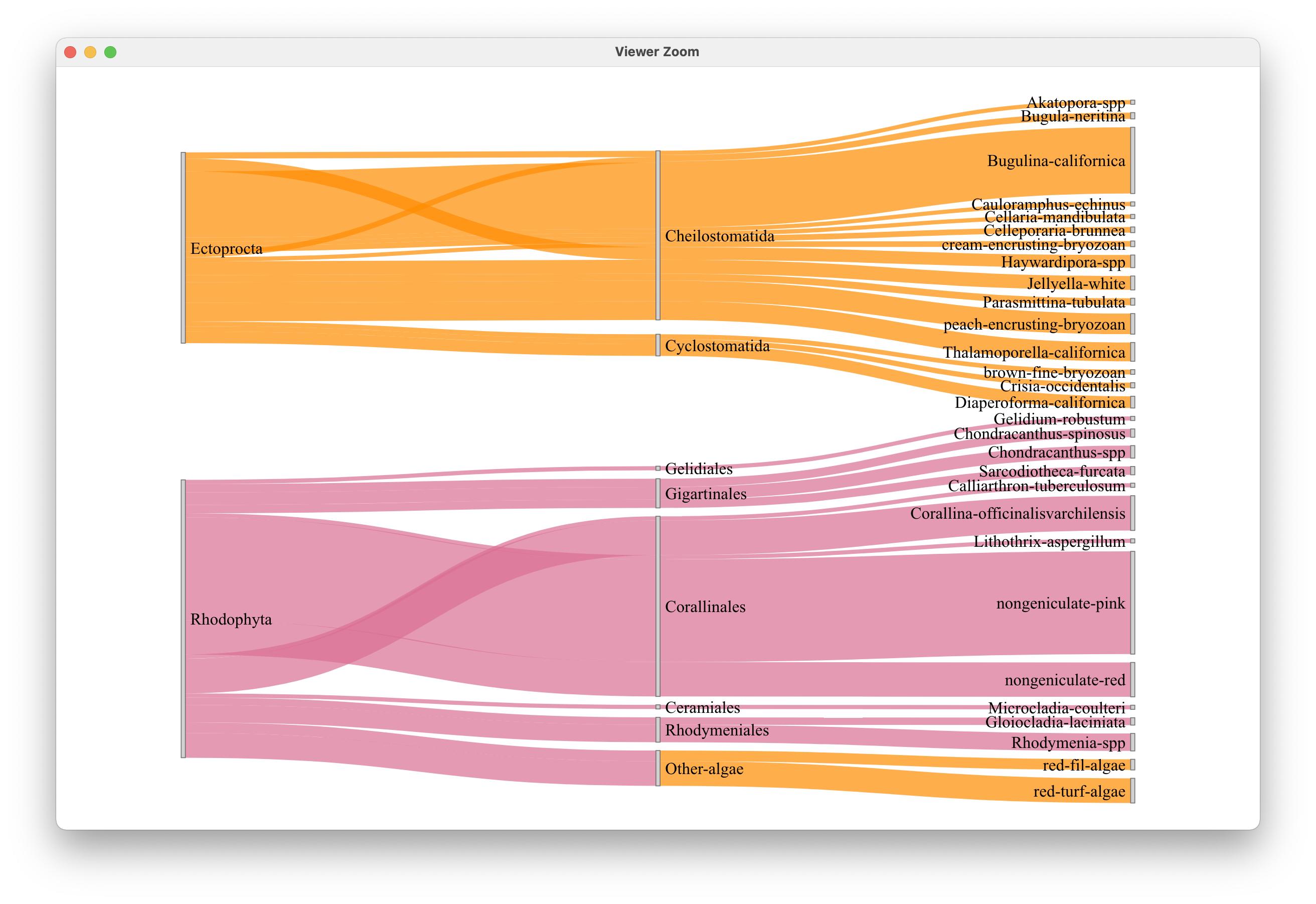
奇怪的是,问题中的链接显示暗橙色-same是顶级类别,而不是像原来的输出那样淡橙色:
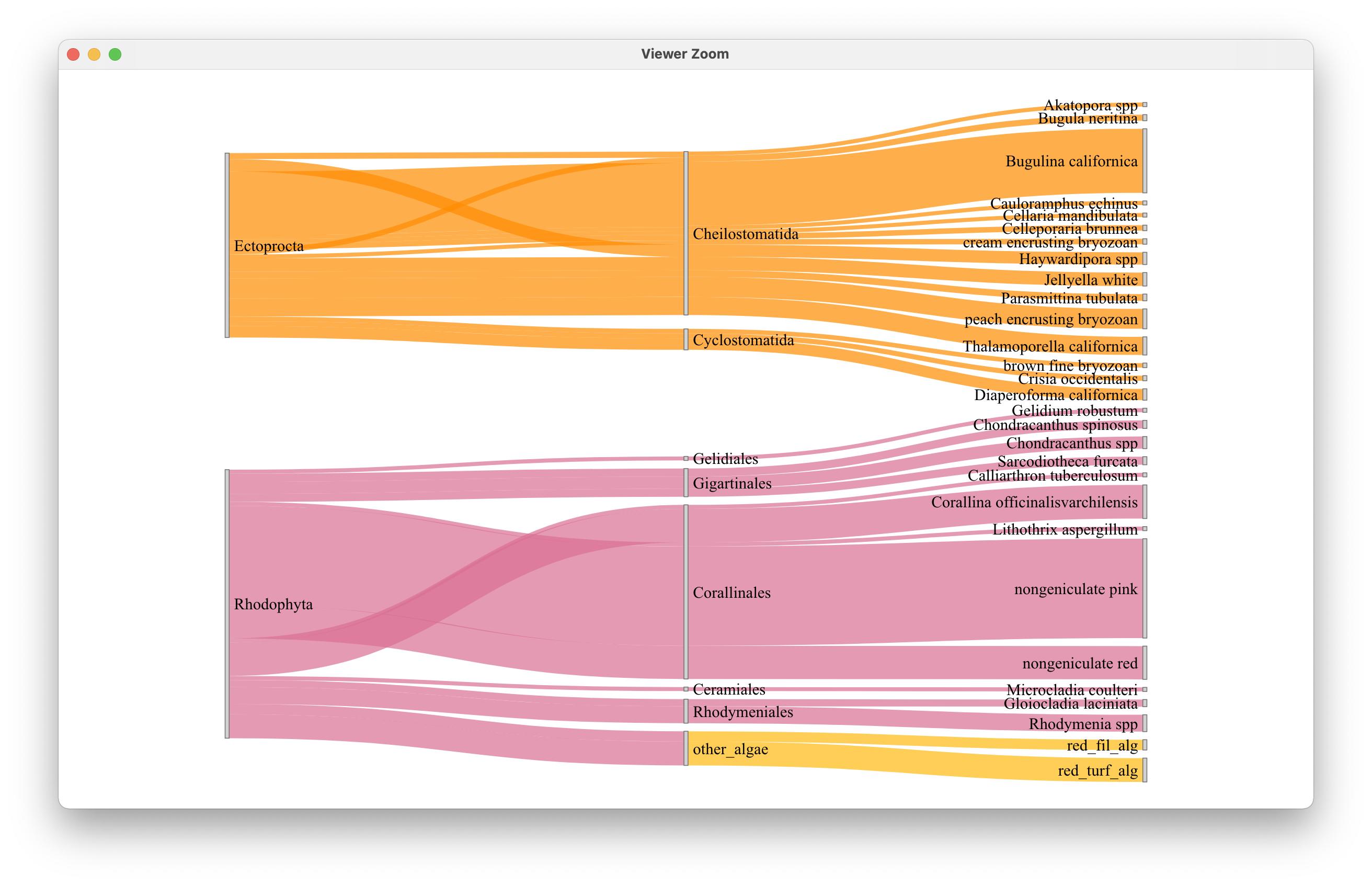
我还尝试了一个标准的D3颜色标度colourScale = JS("d3.scaleOrdinal(d3.schemeCategory20)"),它似乎在原始节点类别中运行得很好:
此外,节点链接结构似乎是正确的,就像构建简单网络(突出显示问题节点)时所看到的那样:
simpleNetwork(links, Source = "source",
Target = "target",
fontSize = 14,
fontFamily = "serif",
linkColour = "#666",
nodeColour = "#69b3a2",
opacity = 0.9,
zoom = T)
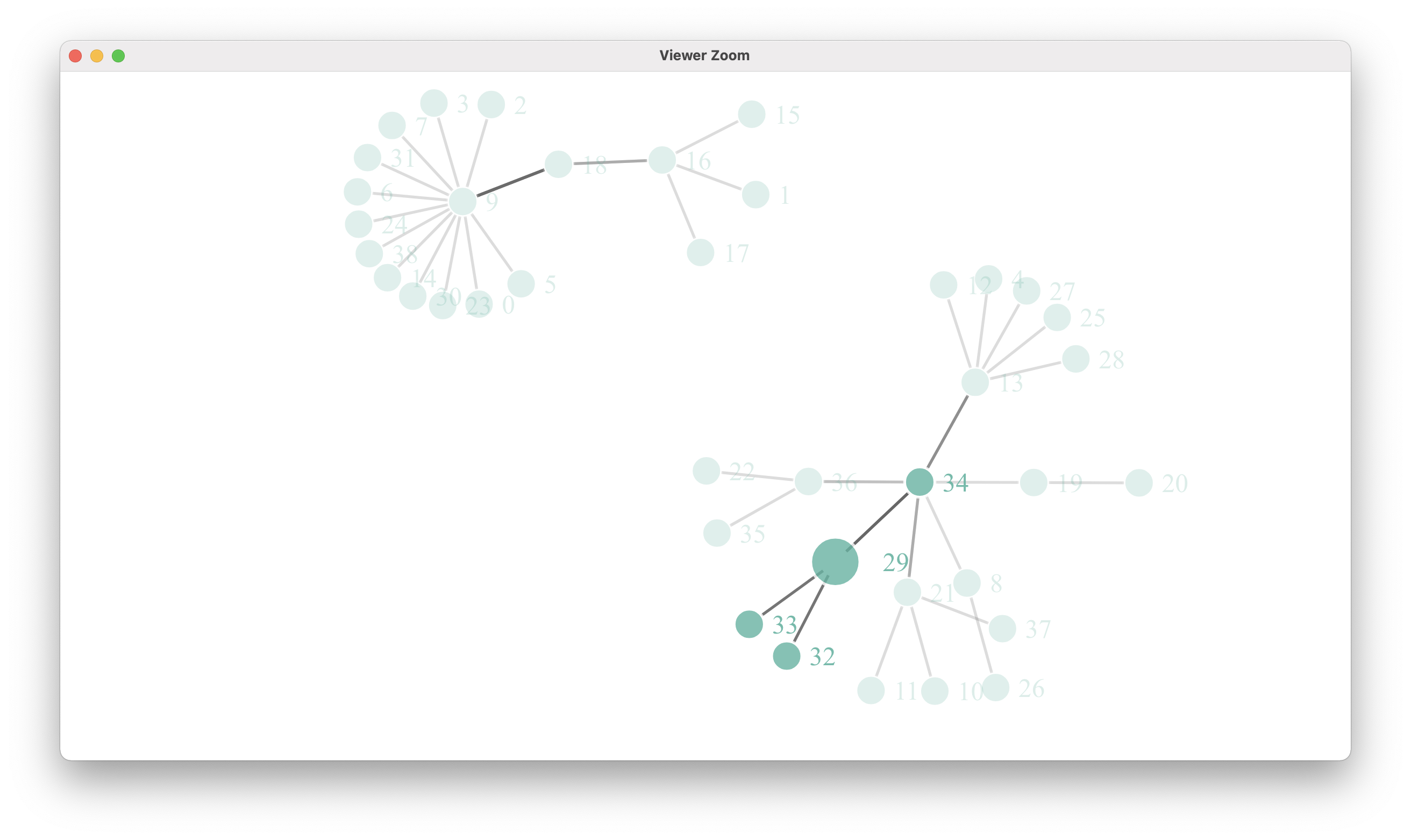
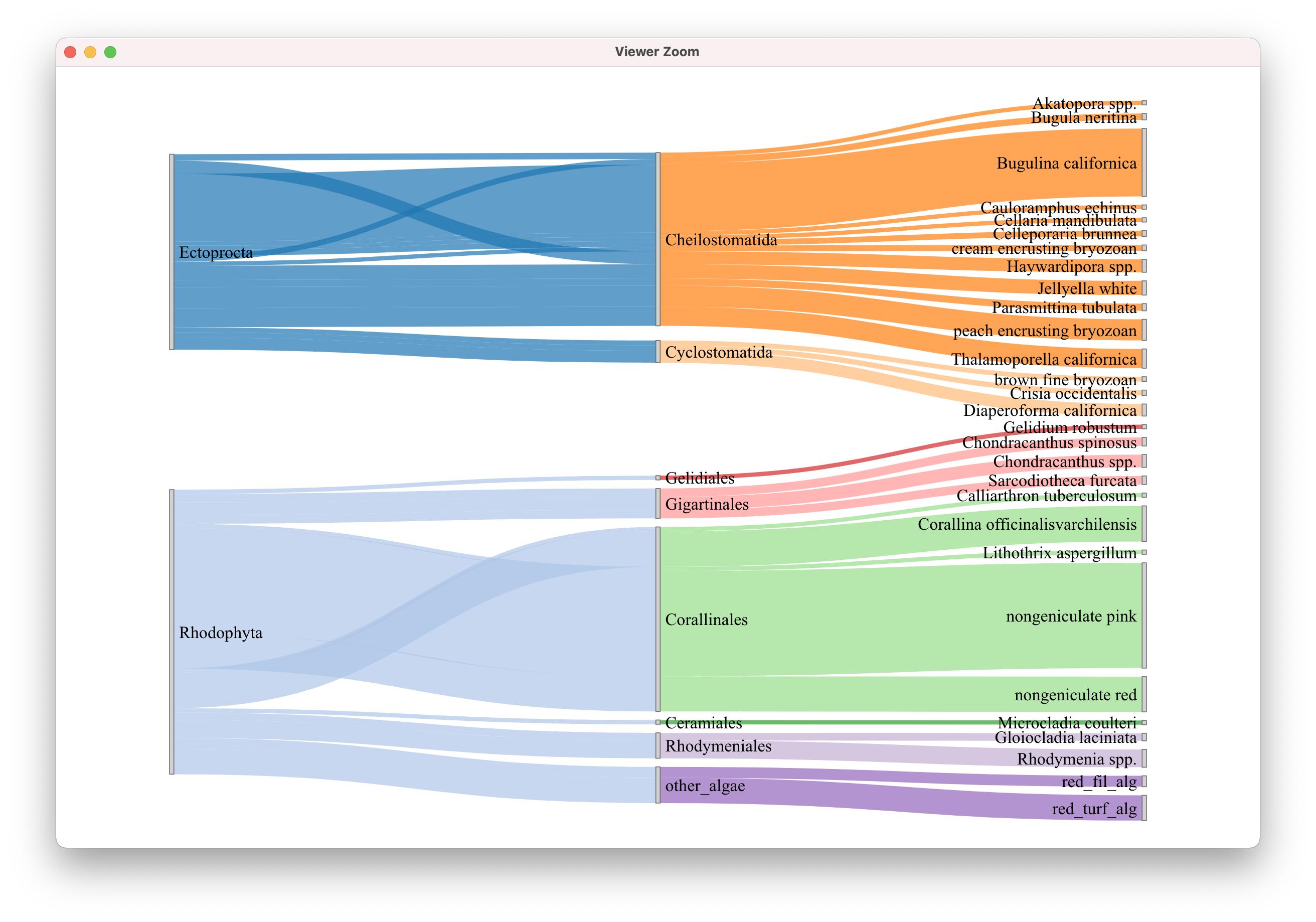
最后,我试图通过一个三色标尺运行域名,以观察其行为。"Ceramiales“类别与”其他藻类“的颜色相同,因此问题与问题类别中的字符串没有直接关系。
colorJS2 <- "d3.scaleOrdinal() .domain([\"Akatopora spp.\", \"brown fine bryozoan\", \"Bugula neritina\", \"Bugulina californica\", \"Calliarthron tuberculosum\", \"Cauloramphus echinus\", \"Cellaria mandibulata\", \"Celleporaria brunnea\", \"Ceramiales\", \"Cheilostomatida\", \"Chondracanthus spinosus\", \"Chondracanthus spp.\", \"Corallina officinalisvarchilensis\", \"Corallinales\", \"cream encrusting bryozoan\", \"Crisia occidentalis\", \"Cyclostomatida\", \"Diaperoforma californica\", \"Ectoprocta\", \"Gelidiales\", \"Gelidium robustum\", \"Gigartinales\", \"Gloiocladia laciniata\", \"Haywardipora spp.\", \"Jellyella white\", \"Lithothrix aspergillum\", \"Microcladia coulteri\", \"nongeniculate pink\", \"nongeniculate red\", \"other_algae\", \"Parasmittina tubulata\", \"peach encrusting bryozoan\", \"red_fil_alg\", \"red_turf_alg\", \"Rhodophyta\", \"Rhodymenia spp.\", \"Rhodymeniales\", \"Sarcodiotheca furcata\", \"Thalamoporella californica\"]) .range([\" #222222\", \"#DB7093\", \"#FFB90F\"])"
sankeyNetwork(Links = links, Nodes = nodes,
Source = "source", Target = "target",
Value = "value", NodeID = "names",
fontSize = 20, nodeWidth = 5,
colourScale = colorJS2,
LinkGroup="group", NodeGroup = NULL)输出:
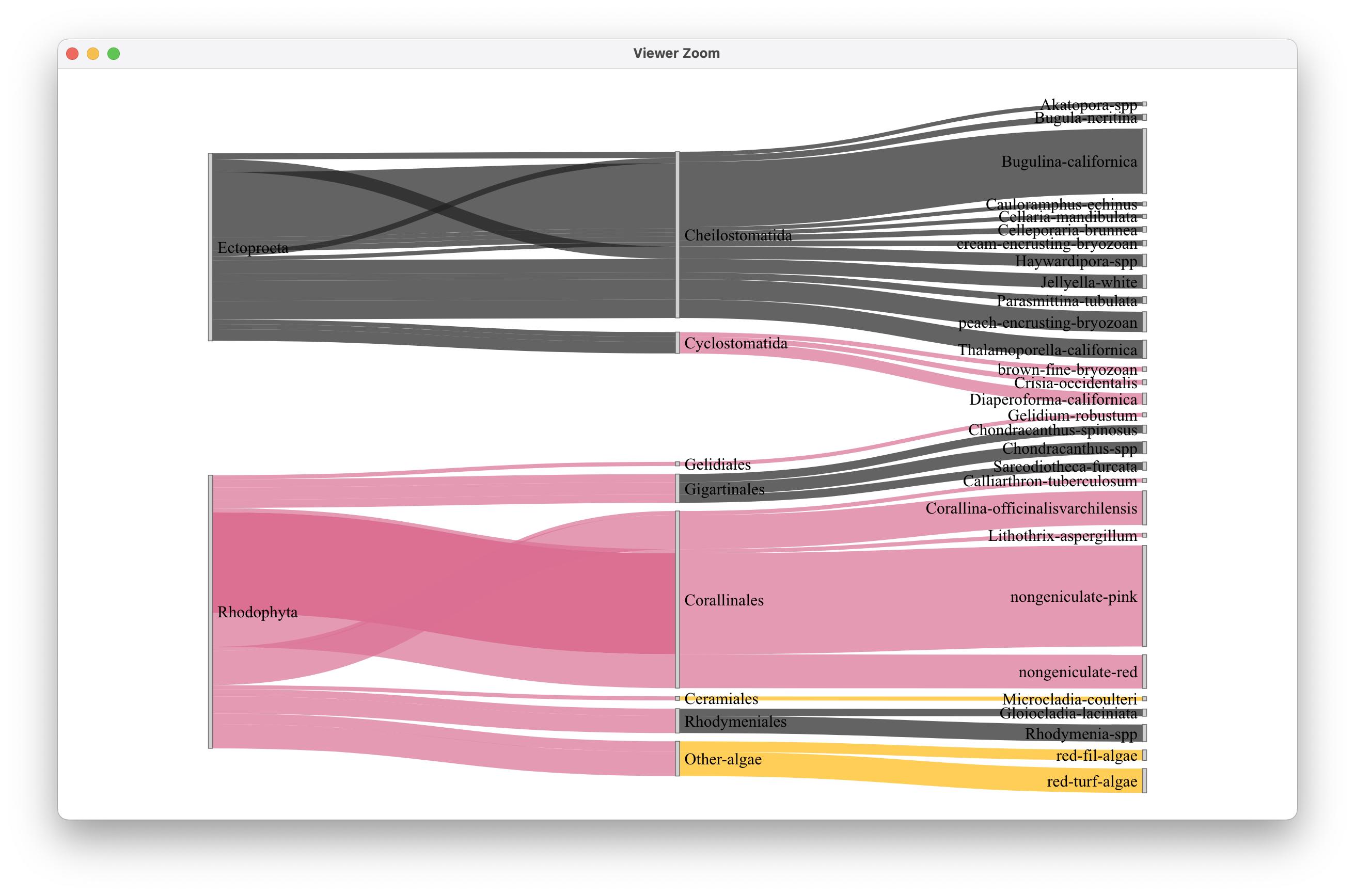
因此,我猜想函数在js域和范围字符串中的运行方式出现了故障。祝好运。
Stack Overflow用户
发布于 2022-01-31 20:17:48
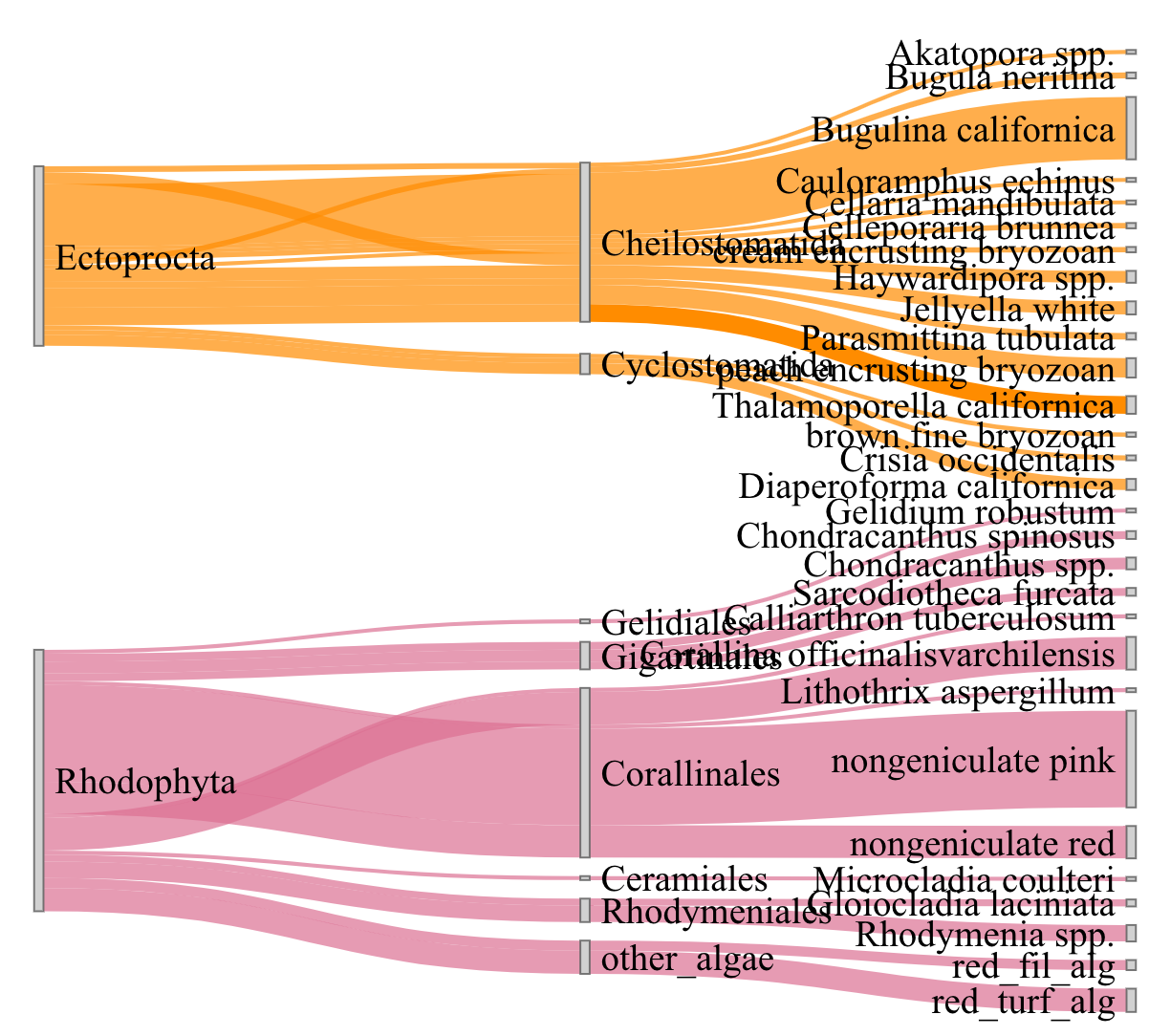
你的颜色列表上有个错误。将"other_algae“的颜色显式设置为"#FFB90F”。如果你修好它,它就会有正确的颜色..。
links <- read.table(header = TRUE, text = "
source target value group
18 9 1.000000 Ectoprocta
18 9 1.500000 Ectoprocta
18 9 15.900000 Ectoprocta
18 9 1.000000 Ectoprocta
18 9 1.000000 Ectoprocta
18 9 1.363636 Ectoprocta
18 9 1.357143 Ectoprocta
18 9 3.025641 Ectoprocta
18 9 3.333333 Ectoprocta
18 9 1.666667 Ectoprocta
18 9 4.934783 Ectoprocta
18 9 4.500000 Ectoprocta
18 16 1.125000 Ectoprocta
18 16 1.250000 Ectoprocta
18 16 2.818182 Ectoprocta
34 8 1.000000 Rhodophyta
34 13 1.000000 Rhodophyta
34 13 8.333333 Rhodophyta
34 13 1.000000 Rhodophyta
34 13 24.655172 Rhodophyta
34 13 8.243902 Rhodophyta
34 19 1.000000 Rhodophyta
34 21 2.000000 Rhodophyta
34 21 3.000000 Rhodophyta
34 21 2.000000 Rhodophyta
34 29 2.555556 Rhodophyta
34 29 5.921569 Rhodophyta
34 36 1.769231 Rhodophyta
34 36 4.187500 Rhodophyta
9 0 1.000000 Cheilostomatida
9 2 1.500000 Cheilostomatida
9 3 15.900000 Cheilostomatida
9 5 1.000000 Cheilostomatida
9 6 1.000000 Cheilostomatida
9 7 1.363636 Cheilostomatida
9 14 1.357143 Cheilostomatida
9 23 3.025641 Cheilostomatida
9 24 3.333333 Cheilostomatida
9 30 1.666667 Cheilostomatida
9 31 4.934783 Cheilostomatida
9 38 4.500000 Cheilostomatida
16 1 1.125000 Cyclostomatida
16 15 1.250000 Cyclostomatida
16 17 2.818182 Cyclostomatida
8 26 1.000000 Ceramiales
13 4 1.000000 Corallinales
13 12 8.333333 Corallinales
13 25 1.000000 Corallinales
13 27 24.655172 Corallinales
13 28 8.243902 Corallinales
19 20 1.000000 Gelidiales
21 10 2.000000 Gigartinales
21 11 3.000000 Gigartinales
21 37 2.000000 Gigartinales
29 32 2.555556 other_algae
29 33 5.921569 other_algae
36 22 1.769231 Rhodymeniales
36 35 4.187500 Rhodymeniales
")
nodes <- read.csv(header = TRUE, text ="
name
Akatopora spp.
brown fine bryozoan
Bugula neritina
Bugulina californica
Calliarthron tuberculosum
Cauloramphus echinus
Cellaria mandibulata
Celleporaria brunnea
Ceramiales
Cheilostomatida
Chondracanthus spinosus
Chondracanthus spp.
Corallina officinalisvarchilensis
Corallinales
cream encrusting bryozoan
Crisia occidentalis
Cyclostomatida
Diaperoforma californica
Ectoprocta
Gelidiales
Gelidium robustum
Gigartinales
Gloiocladia laciniata
Haywardipora spp.
Jellyella white
Lithothrix aspergillum
Microcladia coulteri
nongeniculate pink
nongeniculate red
other_algae
Parasmittina tubulata
peach encrusting bryozoan
red_fil_alg
red_turf_alg
Rhodophyta
Rhodymenia spp.
Rhodymeniales
Sarcodiotheca furcata
Thalamoporella californica
")
colors <- read.csv(header = TRUE, strip.white = TRUE, text = "
domain , range
Akatopora spp. , #FF8C00
brown fine bryozoan , #FF8C00
Bugula neritina , #FF8C00
Bugulina californica , #FF8C00
Calliarthron tuberculosum , #DB7093
Cauloramphus echinus , #FF8C00
Cellaria mandibulata , #FF8C00
Celleporaria brunnea , #FF8C00
Ceramiales , #DB7093
Cheilostomatida , #FF8C00
Chondracanthus spinosus , #DB7093
Chondracanthus spp. , #DB7093
Corallina officinalisvarchilensis , #DB7093
Corallinales , #DB7093
cream encrusting bryozoan , #FF8C00
Crisia occidentalis , #FF8C00
Cyclostomatida , #FF8C00
Diaperoforma californica , #FF8C00
Ectoprocta , #FF8C00
Gelidiales , #DB7093
Gelidium robustum , #DB7093
Gigartinales , #DB7093
Gloiocladia laciniata , #DB7093
Haywardipora spp. , #FF8C00
Jellyella white , #FF8C00
Lithothrix aspergillum , #DB7093
Microcladia coulteri , #DB7093
nongeniculate pink , #DB7093
nongeniculate red , #DB7093
other_algae , #DB7093
Parasmittina tubulata , #FF8C00
peach encrusting bryozoan , #FF8C00
red_fil_alg , #DB7093
red_turf_alg , #DB7093
Rhodophyta , #DB7093
Rhodymenia spp. , #DB7093
Rhodymeniales , #DB7093
Sarcodiotheca furcata , #DB7093
Thalamoporella californica , #FF8C00
")
library(networkD3)
colorJS <- paste0("d3.scaleOrdinal().domain([\"", paste(colors$domain, collapse = "\", \""), "\"]).range([\"", paste(colors$range, collapse = "\", \""), "\"])")
sankeyNetwork(Links = links, Nodes = nodes,
Source = "source", Target = "target",
Value = "value", NodeID = "name",
fontSize = 20, nodeWidth = 5,
colourScale = colorJS,
LinkGroup="group", NodeGroup = NULL)
https://stackoverflow.com/questions/62459949
复制相似问题

