R(二元LISA)中的二元空间相关映射
我想要创建一个地图,显示两个变量之间的二元空间相关性。这可以通过双变量Moran的I空间相关性的LISA映射或使用Lee (2001)提出的L指数来实现。
二元莫兰的I不是在spdep库中实现的,但是L索引是实现的,所以这里是我尝试过的,但没有成功地使用L索引。给出一个基于莫兰的解决方案的答案,我也会非常欢迎!
从下面的可重复示例中可以看到,到目前为止,我一直在计算本地L索引。,我想做的是估计伪p值和创建一个结果的地图,就像我们在LISA空间集群中使用的那样,高-高-低,.,低-低。
在这个例子中,我们的目标是在黑人和白人之间创建一个带有二元Lisa关联的地图。应该在ggplot2中创建地图,显示集群:
- 黑人和白人的高出席率
- 黑人人数多,白人人数少
- 黑人人数少,白人人数多
- 黑人的低存在和低存在哦白人
可复制示例
library(UScensus2000tract)
library(ggplot2)
library(spdep)
library(sf)
# load data
data("oregon.tract")
# plot Census Tract map
plot(oregon.tract)
# Variables to use in the correlation: white and black population in each census track
x <- scale(oregon.tract$white)
y <- scale(oregon.tract$black)
# create Queen contiguity matrix and Spatial weights matrix
nb <- poly2nb(oregon.tract)
lw <- nb2listw(nb)
# Lee index
Lxy <-lee(x, y, lw, length(x), zero.policy=TRUE)
# Lee’s L statistic (Global)
Lxy[1]
#> -0.1865688811
# 10k permutations to estimate pseudo p-values
LMCxy <- lee.mc(x, y, nsim=10000, lw, zero.policy=TRUE, alternative="less")
# quik plot of local L
Lxy[[2]] %>% density() %>% plot() # Lee’s local L statistic (Local)
LMCxy[[7]] %>% density() %>% lines(col="red") # plot values simulated 10k times
# get confidence interval of 95% ( mean +- 2 standard deviations)
two_sd_above <- mean(LMCxy[[7]]) + 2 * sd(LMCxy[[7]])
two_sd_below <- mean(LMCxy[[7]]) - 2 * sd(LMCxy[[7]])
# convert spatial object to sf class for easier/faster use
oregon_sf <- st_as_sf(oregon.tract)
# add L index values to map object
oregon_sf$Lindex <- Lxy[[2]]
# identify significant local results
oregon_sf$sig <- if_else( oregon_sf$Lindex < 2*two_sd_below, 1, if_else( oregon_sf$Lindex > 2*two_sd_above, 1, 0))
# Map of Local L index but only the significant results
ggplot() + geom_sf(data=oregon_sf, aes(fill=ifelse( sig==T, Lindex, NA)), color=NA)

回答 3
Stack Overflow用户
发布于 2017-07-23 14:53:38
那这个呢?
我用的是普通的莫兰指数,而不是你建议的李指数。但我认为基本的推理是一样的。
你可能会看到下面的结果--以这种方式产生的结果非常类似于从GeoDA开始的结果。
library(dplyr)
library(ggplot2)
library(sf)
library(spdep)
library(rgdal)
library(stringr)
library(UScensus2000tract)
#======================================================
# load data
data("oregon.tract")
# Variables to use in the correlation: white and black population in each census track
x <- oregon.tract$white
y <- oregon.tract$black
#======================================================
# Programming some functions
# Bivariate Moran's I
moran_I <- function(x, y = NULL, W){
if(is.null(y)) y = x
xp <- (x - mean(x, na.rm=T))/sd(x, na.rm=T)
yp <- (y - mean(y, na.rm=T))/sd(y, na.rm=T)
W[which(is.na(W))] <- 0
n <- nrow(W)
global <- (xp%*%W%*%yp)/(n - 1)
local <- (xp*W%*%yp)
list(global = global, local = as.numeric(local))
}
# Permutations for the Bivariate Moran's I
simula_moran <- function(x, y = NULL, W, nsims = 1000){
if(is.null(y)) y = x
n = nrow(W)
IDs = 1:n
xp <- (x - mean(x, na.rm=T))/sd(x, na.rm=T)
W[which(is.na(W))] <- 0
global_sims = NULL
local_sims = matrix(NA, nrow = n, ncol=nsims)
ID_sample = sample(IDs, size = n*nsims, replace = T)
y_s = y[ID_sample]
y_s = matrix(y_s, nrow = n, ncol = nsims)
y_s <- (y_s - apply(y_s, 1, mean))/apply(y_s, 1, sd)
global_sims <- as.numeric( (xp%*%W%*%y_s)/(n - 1) )
local_sims <- (xp*W%*%y_s)
list(global_sims = global_sims,
local_sims = local_sims)
}
#======================================================
# Adjacency Matrix (Queen)
nb <- poly2nb(oregon.tract)
lw <- nb2listw(nb, style = "B", zero.policy = T)
W <- as(lw, "symmetricMatrix")
W <- as.matrix(W/rowSums(W))
W[which(is.na(W))] <- 0
#======================================================
# Calculating the index and its simulated distribution
# for global and local values
m <- moran_I(x, y, W)
m[[1]] # global value
m_i <- m[[2]] # local values
local_sims <- simula_moran(x, y, W)$local_sims
# Identifying the significant values
alpha <- .05 # for a 95% confidence interval
probs <- c(alpha/2, 1-alpha/2)
intervals <- t( apply(local_sims, 1, function(x) quantile(x, probs=probs)))
sig <- ( m_i < intervals[,1] ) | ( m_i > intervals[,2] )
#======================================================
# Preparing for plotting
oregon.tract <- st_as_sf(oregon.tract)
oregon.tract$sig <- sig
# Identifying the LISA patterns
xp <- (x-mean(x))/sd(x)
yp <- (y-mean(y))/sd(y)
patterns <- as.character( interaction(xp > 0, W%*%yp > 0) )
patterns <- patterns %>%
str_replace_all("TRUE","High") %>%
str_replace_all("FALSE","Low")
patterns[oregon.tract$sig==0] <- "Not significant"
oregon.tract$patterns <- patterns
# Plotting
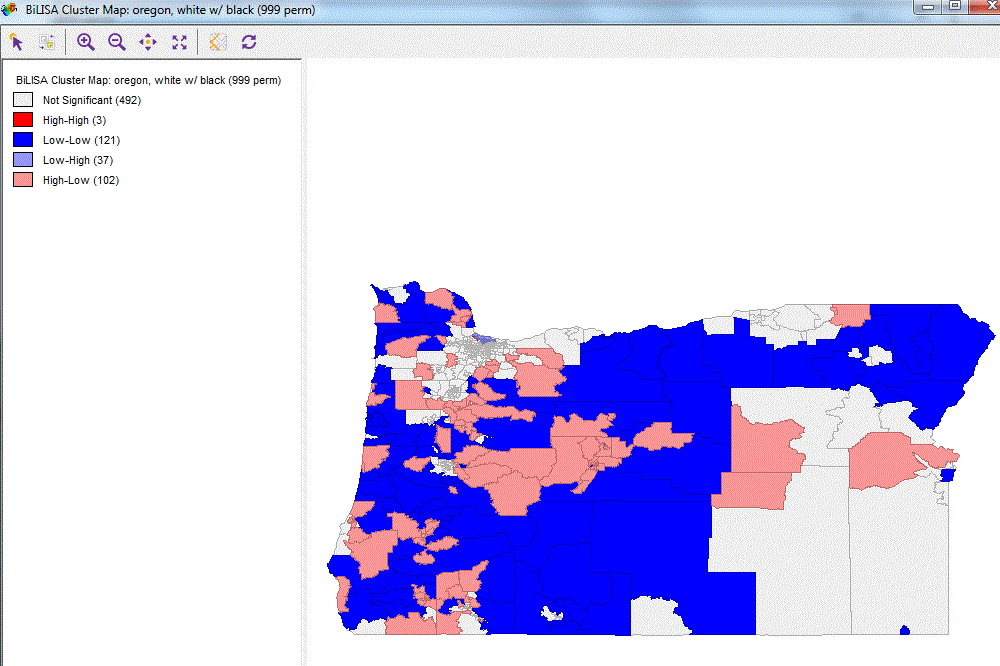
ggplot() + geom_sf(data=oregon.tract, aes(fill=patterns), color="NA") +
scale_fill_manual(values = c("red", "pink", "light blue", "dark blue", "grey95")) +
theme_minimal()通过对置信区间进行更改(例如使用90%而不是95%),可以使结果更接近(但不完全相同) GeoDa的结果。
我认为其余的差异来自于计算Moran's I的稍微不同的方法。我的版本给出了包moran中可用的函数spdep的相同值。但是GeoDa可能会使用另一种方法。
Stack Overflow用户
发布于 2020-11-05 21:28:42
我想现在添加到线程中已经很晚了,但是Lee的L与您在这里所做的完全不同,这是Wartenberg (1985)的创新。这有一些潜在的缺点。主要研究了x与y作为@RogerioJB的滞后之间的关系,阐明了空间滞后y是通过将模拟的y乘以邻接矩阵来计算的。Lee (2001)的创新是完全不同的,涉及到Pearson的r和空间平滑标量(SSS)的集成,而是比较了x和y之间的过程,而不是y的滞后。采用的@RogerioJB方法可以通过从lee.mc函数生成可能的局部l分布来复制。反过来,结果可以用类似于GeoDa的高高的样式绘制.低显着性聚类图。
Stack Overflow用户
发布于 2020-12-06 07:13:54
在@justin-k的建议的基础上,我修改了双变量的LISA代码,通过@rogeriojb来计算Lee的L统计量。此方法从lee.mc()包中创建一个修改后的斯戴普函数,以模拟本地L值。我在具有点级数据的GitHub要点中提供了另一个示例。
library(boot)
library(dplyr)
library(ggplot2)
library(sf)
library(spdep)
library(rgdal)
library(stringr)
library(UScensus2000tract)
#======================================================
# load data
data("oregon.tract")
# Variables to use in the correlation: white and black population in each census track
x <- oregon.tract$white
y <- oregon.tract$black
# ----------------------------------------------------- #
# Program a function
## Permutations for Lee's L statistic
## Modification of the lee.mc() function within the {spdep} package
## Saves 'localL' output instead of 'L' output
simula_lee <- function(x, y, listw, nsim = nsim, zero.policy = NULL, na.action = na.fail) {
if (deparse(substitute(na.action)) == "na.pass")
stop ("na.pass not permitted")
na.act <- attr(na.action(cbind(x, y)), "na.action")
x[na.act] <- NA
y[na.act] <- NA
x <- na.action(x)
y <- na.action(y)
if (!is.null(na.act)) {
subset <- !(1:length(listw$neighbours) %in% na.act)
listw <- subset(listw, subset, zero.policy = zero.policy)
}
n <- length(listw$neighbours)
if ((n != length(x)) | (n != length(y)))
stop ("objects of different length")
gamres <- suppressWarnings(nsim > gamma(n + 1))
if (gamres)
stop ("nsim too large for this number of observations")
if (nsim < 1)
stop ("nsim too small")
xy <- data.frame(x, y)
S2 <- sum((unlist(lapply(listw$weights, sum)))^2)
lee_boot <- function(var, i, ...) {
return(lee(x = var[i, 1], y = var[i, 2], ...)$localL)
}
res <- boot(xy, statistic = lee_boot, R = nsim, sim = "permutation",
listw = listw, n = n, S2 = S2, zero.policy = zero.policy)
}
# ----------------------------------------------------- #
# Adjacency Matrix
nb <- poly2nb(oregon.tract)
lw <- nb2listw(nb, style = "B", zero.policy = T)
W <- as(lw, "symmetricMatrix")
W <- as.matrix(W / rowSums(W))
W[which(is.na(W))] <- 0
# ----------------------------------------------------- #
# Calculate the index and its simulated distribution
# for global and local values
# Global Lee's L
lee.test(x = x, y = y, listw = lw, zero.policy = TRUE,
alternative = "two.sided", na.action = na.omit)
# Local Lee's L values
m <- lee(x = x, y = y, listw = lw, n = length(x),
zero.policy = TRUE, NAOK = TRUE)
# Local Lee's L simulations
local_sims <- simula_lee(x = x, y = y, listw = lw, nsim = 10000,
zero.policy = TRUE, na.action = na.omit)
m_i <- m[[2]] # local values
# Identify the significant values
alpha <- 0.05 # for a 95% confidence interval
probs <- c(alpha/2, 1-alpha/2)
intervals <- t(apply(t(local_sims[[2]]), 1, function(x) quantile(x, probs = probs)))
sig <- (m_i < intervals[ , 1] ) | ( m_i > intervals[ , 2])
#======================================================
# Preparing for plotting
oregon.tract <- st_as_sf(oregon.tract)
oregon.tract$sig <- sig
# Identifying the Lee's L patterns
xp <- scale(x)
yp <- scale(y)
patterns <- as.character(interaction(xp > 0, W%*%yp > 0))
patterns <- patterns %>%
str_replace_all("TRUE","High") %>%
str_replace_all("FALSE","Low")
patterns[oregon.tract$sig == 0] <- "Not significant"
oregon.tract$patterns <- patterns
# Plotting
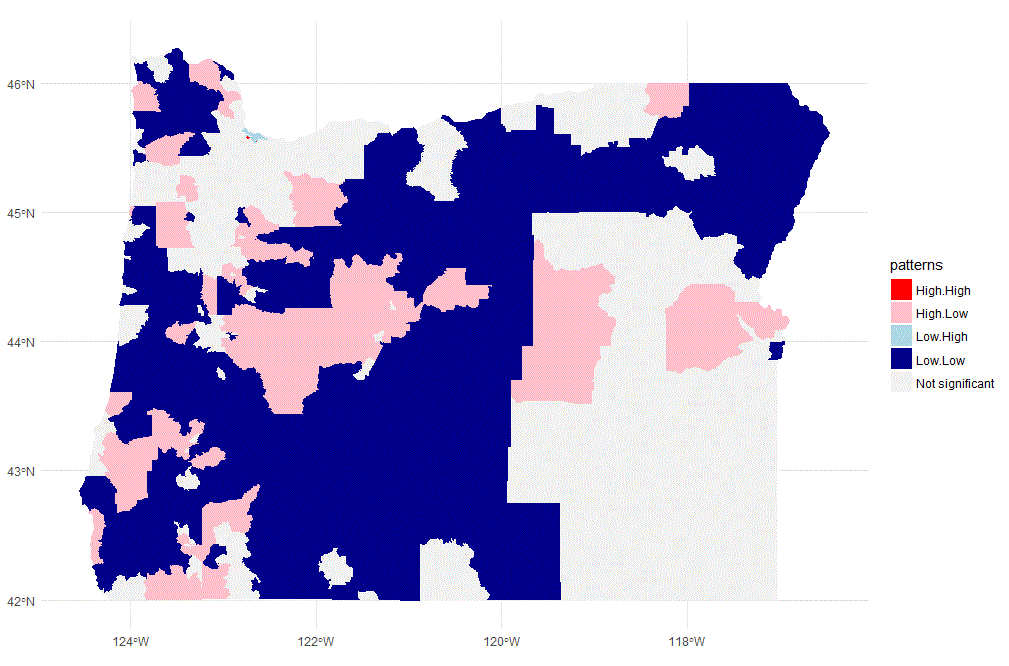
ggplot() +
geom_sf(data = oregon.tract, aes(fill = patterns), color = "NA") +
scale_fill_manual(values = c("red", "pink", "light blue", "dark blue", "grey95")) +
guides(fill = guide_legend(title = "Lee's L clusters")) +
theme_minimal()https://stackoverflow.com/questions/45177590
复制相似问题

