在R中标出一个谱系
在R中标出一个谱系
提问于 2018-02-26 05:13:32
d = data.frame(
offspring = c("G2I1", "G2I2", "G2I3", "G3I1", "G3I2", "G3I3", "G3I4", "G4I1", "G4I2", "G4I3", "G4I4", "G5I1", "G5I2", "G5I3" ),
parent1 = c("G1I1", "G1I2", "G1I1", "G2I1", "G2I3", "G2I1", "G2I3", "G3I2", "G3I2", "G3I1", "G3I4", "G4I3", "G4I3", "G4I1" ),
parent2 = c("G1I3", "G1I2", "G1I2", "G2I2", "G2I2", "G2I2", "G2I3", "G3I4", "G3I1", "G3I2", "G3I4", "G4I1", "G4I1", "G4I2" )
)
print(d)
offspring parent1 parent2
1 G2I1 G1I1 G1I3 # generation 2
2 G2I2 G1I2 G1I2 # generation 2
3 G2I3 G1I1 G1I2 # generation 2
4 G3I1 G2I1 G2I2 # generation 3
5 G3I2 G2I3 G2I2 # generation 3
6 G3I3 G2I1 G2I2 # generation 3
7 G3I4 G2I3 G2I3 # generation 3
8 G4I1 G3I2 G3I4 # generation 4
9 G4I2 G3I2 G3I1 # generation 4
10 G4I3 G3I1 G3I2 # generation 4
11 G4I4 G3I4 G3I4 # generation 4
12 G5I1 G4I3 G4I1 # generation 5
13 G5I2 G4I3 G4I1 # generation 5
14 G5I3 G4I1 G4I2 # generation 5数据表示
此数据表示家谱。每一行表示一个后代及其双亲。我称他们为parent1和parent2,因为他们是两性人。而且,他们也可以克隆自己!世代是不重叠的,这意味着n一代的所有父母都是在n-1世代出生的。
让我们考虑一个例子。个人G3I4诞生于第三代(G3),是这一代的个体索引4 (I4;索引只是一个ID)。该个体是个体G4I1和个体G4I4的父个体。事实上,G3I4是G4I4克隆自己的唯一父母。
问题
我如何在R中绘制这个家谱?
相关员额
post How to plot family tree in R是非常相关的,但是我没有将它应用到我的数据中。第一个问题使用的是我不太熟悉的igraph。但我没能得到任何好看的东西
d = tibble(
offspring = c("G2I1", "G2I2", "G2I3", "G3I1", "G3I2", "G3I3", "G3I4", "G4I1", "G4I2", "G4I3", "G4I4", "G5I1", "G5I2", "G5I3" ),
parent1 = c("G1I1", "G1I2", "G1I1", "G2I1", "G2I3", "G2I1", "G2I3", "G3I2", "G3I2", "G3I1", "G3I4", "G4I3", "G4I3", "G4I1" ),
parent2 = c("G1I3", "G1I2", "G1I2", "G2I2", "G2I2", "G2I2", "G2I3", "G3I4", "G3I1", "G3I2", "G3I4", "G4I1", "G4I1", "G4I2" )
)
d2 = data.frame(from=c(d$parent1,d$parent2), to=rep(d$offspring,2))
g=graph_from_data_frame(d2)
co=layout.reingold.tilford(g, flip.y=T)
plot(g,layout=co)
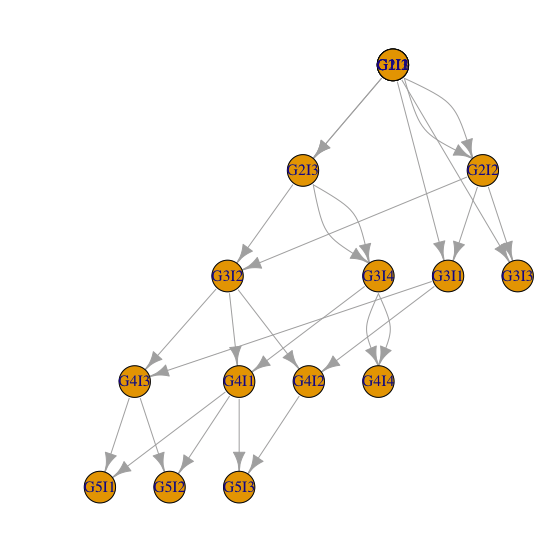
但是一些没有留下任何后代的个体却从图表中消失了。
第二个答案使用kinship2。据我所知,kinship2不能处理无性繁殖。
回答 1
Stack Overflow用户
回答已采纳
发布于 2018-02-28 21:34:59
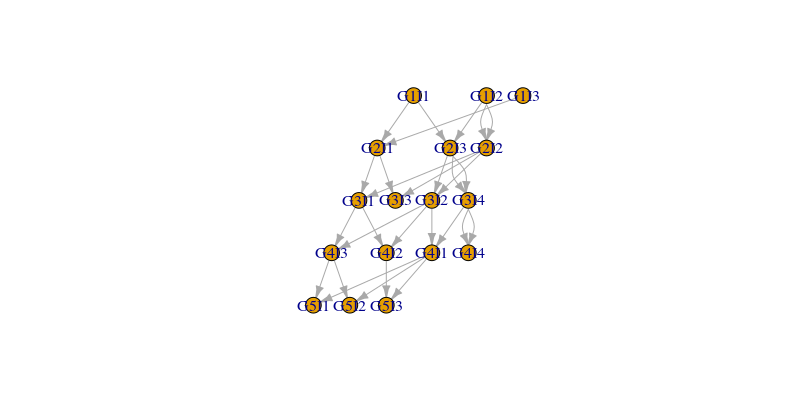
我看到的唯一错误是G1中的重叠节点。有了更多的信息,我很乐意在必要时调整输出。
library(igraph)
d = data.frame(
offspring = c("G2I1", "G2I2", "G2I3", "G3I1", "G3I2", "G3I3", "G3I4", "G4I1", "G4I2", "G4I3", "G4I4", "G5I1", "G5I2", "G5I3" ),
parent1 = c("G1I1", "G1I2", "G1I1", "G2I1", "G2I3", "G2I1", "G2I3", "G3I2", "G3I2", "G3I1", "G3I4", "G4I3", "G4I3", "G4I1" ),
parent2 = c("G1I3", "G1I2", "G1I2", "G2I2", "G2I2", "G2I2", "G2I3", "G3I4", "G3I1", "G3I2", "G3I4", "G4I1", "G4I1", "G4I2" ),
stringsAsFactors = F
)
d2 = data.frame(from=c(d$parent1,d$parent2), to=rep(d$offspring,2))
g=graph_from_data_frame(d2)
#co=layout.reingold.tilford(g, flip.y=T)
co1 <- layout_as_tree(g, root = which(grepl("G1", V(g)$name)))
#plot(g,layout=co, edge.arrow.size=0.5)
plot(g,layout=co1, edge.arrow.size=0.25)
页面原文内容由Stack Overflow提供。腾讯云小微IT领域专用引擎提供翻译支持
原文链接:
https://stackoverflow.com/questions/48981822
复制相关文章
相似问题

