使用“nVennR”包转换数据以创建广义的准比例Venn图
我有下面的数据集,并希望您帮助转换它,以便能够使用包装‘nVennR’ by Pérez-Silva等人。2018年绘制Venn。
以下是数据集:
dput(data)
structure(list(Employee = c("A001", "A002", "A003", "A004", "A005",
"A006", "A007", "A008", "A009", "A010", "A011", "A012", "A013",
"A014", "A015", "A016", "A017", "A018"), SAS = c("Y", "N", "Y",
"Y", "Y", "Y", "N", "Y", "N", "N", "Y", "Y", "Y", "Y", "N", "N",
"N", "N"), Python = c("Y", "Y", "Y", "Y", "N", "N", "N", "N",
"N", "N", "Y", "Y", "N", "N", "N", "N", "Y", "Y"), R = c("Y",
"Y", "N", "Y", "N", "Y", "N", "N", "Y", "Y", "Y", "Y", "Y", "Y",
"Y", "Y", "N", "N")), .Names = c("Employee", "SAS", "Python",
"R"), row.names = c(NA, -18L), class = c("tbl_df", "tbl", "data.frame"
))下面是我想要得到的Venn图的一个例子:
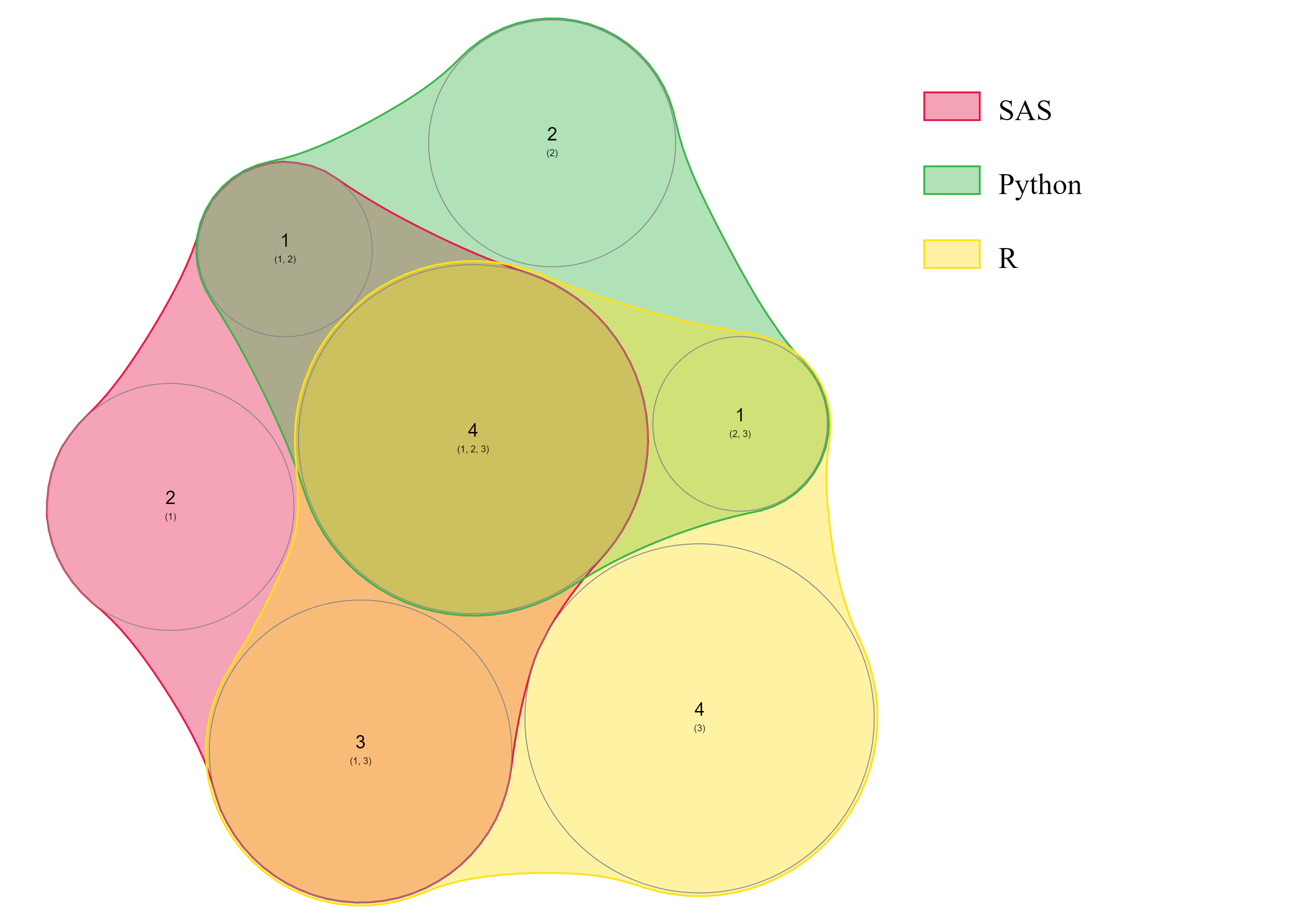
更新:
在安装nVennR和rsvg的更新版本之后,当我从这里运行示例代码时,会得到下面的错误和图表:
Warning message:
In checkValidSVG(doc, warn = warn) :
This picture was not generated by the 'grConvert' package, errors may result
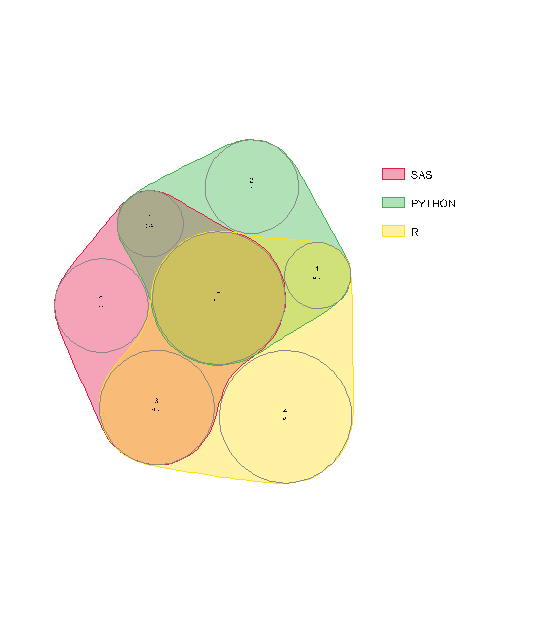
下面是我的会议信息:
sessionInfo()
R version 3.4.2 (2017-09-28)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows >= 8 x64 (build 9200)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods
[7] base
other attached packages:
[1] nVennR_0.2.0
loaded via a namespace (and not attached):
[1] Rcpp_0.12.16 lattice_0.20-35 XML_3.98-1.10
[4] png_0.1-7 rsvg_1.1 grid_3.4.2
[7] plyr_1.8.4 gtable_0.2.0 scales_0.5.0.9000
[10] ggplot2_2.2.1.9000 pillar_1.2.1 rlang_0.2.0.9001
[13] grImport2_0.1-2 lazyeval_0.2.1 Matrix_1.2-12
[16] tools_3.4.2 munsell_0.4.3 jpeg_0.1-8
[19] compiler_3.4.2 base64enc_0.1-3 colorspace_1.3-2
[22] tibble_1.4.2如果有任何解决这个问题的想法,我将不胜感激。
回答 4
Stack Overflow用户
发布于 2018-04-16 10:05:37
让您知道nVennR的新版本已经准备好了。现在输入和输出控制是不同的,toVenn被废弃了,由plotVenn代替。有一个有几个例子的小片段,其中一个使用了这个问题中的数据,这里。
Stack Overflow用户
发布于 2018-03-25 02:11:01
下面是一种在Bioconductor中使用Bioconductor包的方法,将您从dput加载的数据作为变量z使用
source("http://www.bioconductor.org/biocLite.R")
biocLite("limma")
library(limma)将“全Y”改为“真”,“全N”改为“假”:
z2 <- data.frame(lapply(z, function(x) { gsub("Y", "TRUE", x) }))
z3 <- data.frame(lapply(z2, function(x) { gsub("N", "FALSE", x) }),stringsAsFactors=FALSE)确保它们都是逻辑类型:
z3$SAS <- as.logical(z3$SAS)
z3$Python <- as.logical(z3$Python)
z3$R <- as.logical(z3$R)现在使用vennCounts汇总每个Venn区域的所有总数。
> ( venn.totals <- vennCounts(z3[,-1]) )
SAS Python R Counts
1 0 0 0 1
2 0 0 1 4
3 0 1 0 2
4 0 1 1 1
5 1 0 0 2
6 1 0 1 3
7 1 1 0 1
8 1 1 1 4
attr(,"class")
[1] "VennCounts"生成图表只是另一个步骤:
vennDiagram(venn.totals)
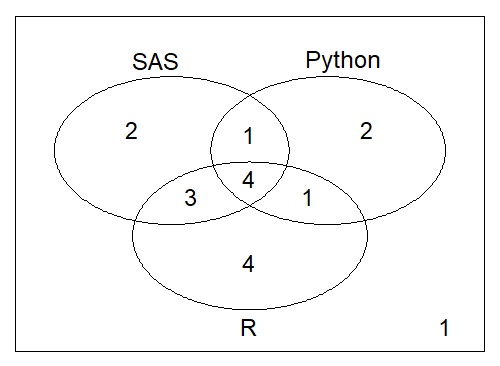
Stack Overflow用户
发布于 2018-03-28 15:43:56
很高兴有这么快的反馈。也许我们应该在文档化中声明这个版本的nVennR是初步的。一些研究人员提出了一种快速运行nVenn的方法,所以我只是把C++代码转换成了几个R函数。如您所见,结果显示在viewer窗口中,而不是plot窗口中。我边走边学。因为我看到了这个包的一些兴趣,所以我正在编译一个特性列表以添加到下一个版本中。更好的输入选项肯定在列表中。另外,对输出进行更多的控制(顺便说一句,如果颜色在路上,您只需将opacity设置为0)。
关于这个问题,“神秘是对的,你会向函数发送列表。”一种快速的方法就是
sas <- subset(data, SAS == "Y")$Employee
python <- subset(data, Python == "Y")$Employee
rr <- subset(data, R == "Y")$Employee
mySVG <- toVenn(sas, python, rr)
showSVG(mySVG = mySVG, opacity = 0.1)下一个版本将有一个单独输入名称的方法(对不起)
https://stackoverflow.com/questions/49471565
复制相似问题
