生信技能树笔记day3
原创title: "生信技能树学习笔记"
引用自生信技能树 author: "天空"
date: "2023-01-02"
output: html_document
一、数据框、矩阵和列表
1. 数据框来源
(1)代码新建数据框
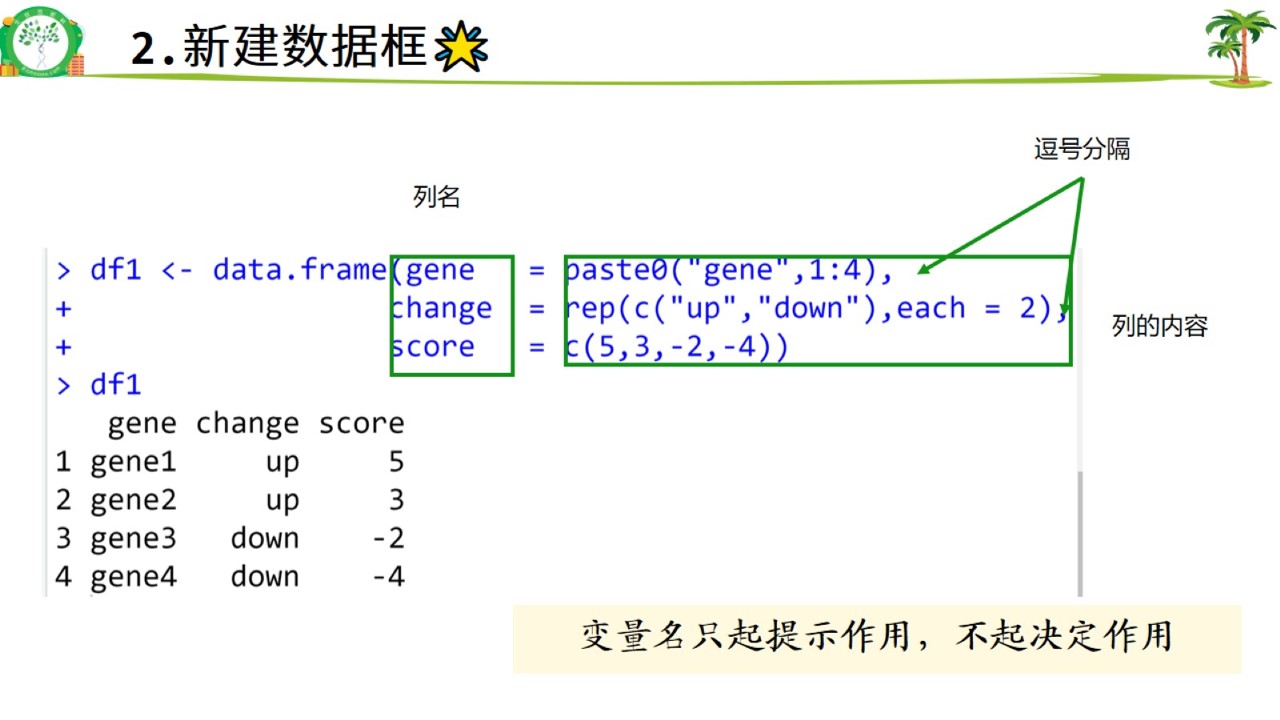
代码新建.jpg
#新建和读取数据框
df1 <- data.frame(gene = paste0("gene",1:4),
change = rep(c("up","down"),each = 2),
score = c(5,3,-2,-4))
df1## gene change score
## 1 gene1 up 5
## 2 gene2 up 3
## 3 gene3 down -2
## 4 gene4 down -4(2)文件中读取
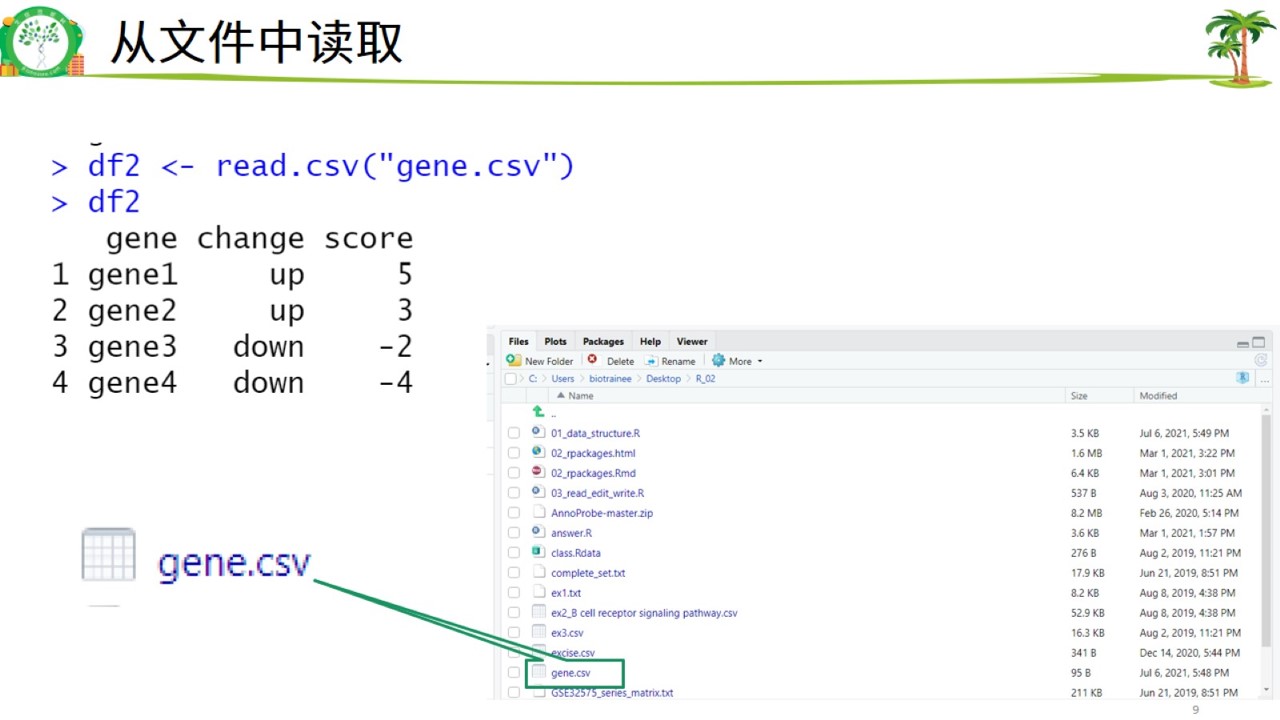
文件读取数据.jpg
df2 <- read.csv("day3/R_02/gene.csv")
df2## gene change score
## 1 gene1 up 5
## 2 gene2 up 3
## 3 gene3 down -2
## 4 gene4 down -42. 数据框属性

数据框属性.jpg
#数据框属性
dim(df1)## [1] 4 3nrow(df1)## [1] 4ncol(df1)## [1] 3rownames(df1)## [1] "1" "2" "3" "4"colnames(df1)## [1] "gene" "change" "score"3. 数据框取子集
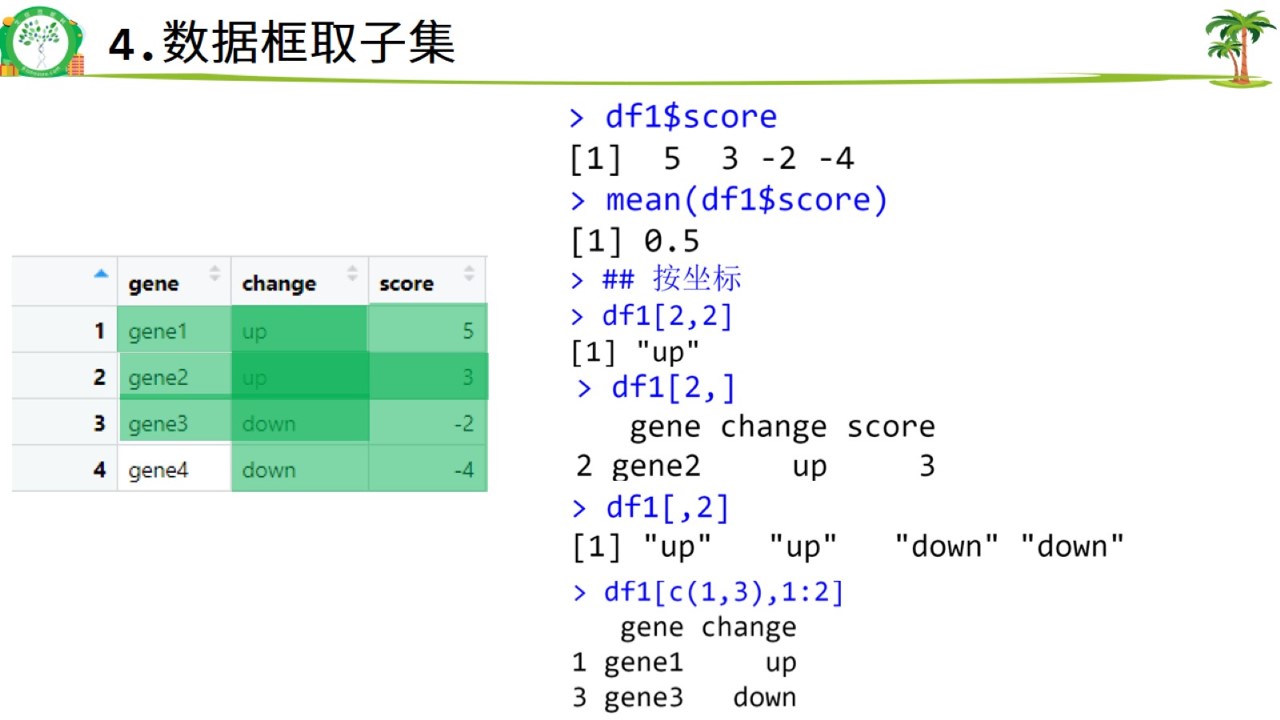
数据框取子集.jpg
#数据框取子集
df1$gene #删掉score,按tab键试试## [1] "gene1" "gene2" "gene3" "gene4"mean(df1$score)## [1] 0.5# 按坐标
df1[2,2]## [1] "up"df1[2,]## gene change score
## 2 gene2 up 3df1[,2]## [1] "up" "up" "down" "down"df1[c(1,3),1:2]## gene change
## 1 gene1 up
## 3 gene3 down# 按名字
df1[,"gene"]## [1] "gene1" "gene2" "gene3" "gene4"typeof(df1[,"gene"])## [1] "character"df1["gene"]## gene
## 1 gene1
## 2 gene2
## 3 gene3
## 4 gene4typeof(df1["gene"])## [1] "list"df1[,c('gene','change')]## gene change
## 1 gene1 up
## 2 gene2 up
## 3 gene3 down
## 4 gene4 down# 按条件(逻辑值)
df1[df1$score>0,]## gene change score
## 1 gene1 up 5
## 2 gene2 up 3# 中括号里面的逗号表示维度分割!!!
# 数据框按照逻辑值取子集,TRUE对应的行/列留下,FALSE对应的行/列丢掉4. 代码思维

代码思维1.jpg
# 代码思维
#如何取数据框的最后一列?
df1[,3]## [1] 5 3 -2 -4df1[,ncol(df1)]## [1] 5 3 -2 -4#如何取数据框除了最后一列以外的其他列?
df1[,-ncol(df1)]## gene change
## 1 gene1 up
## 2 gene2 up
## 3 gene3 down
## 4 gene4 down5. 数据框修改行/列名
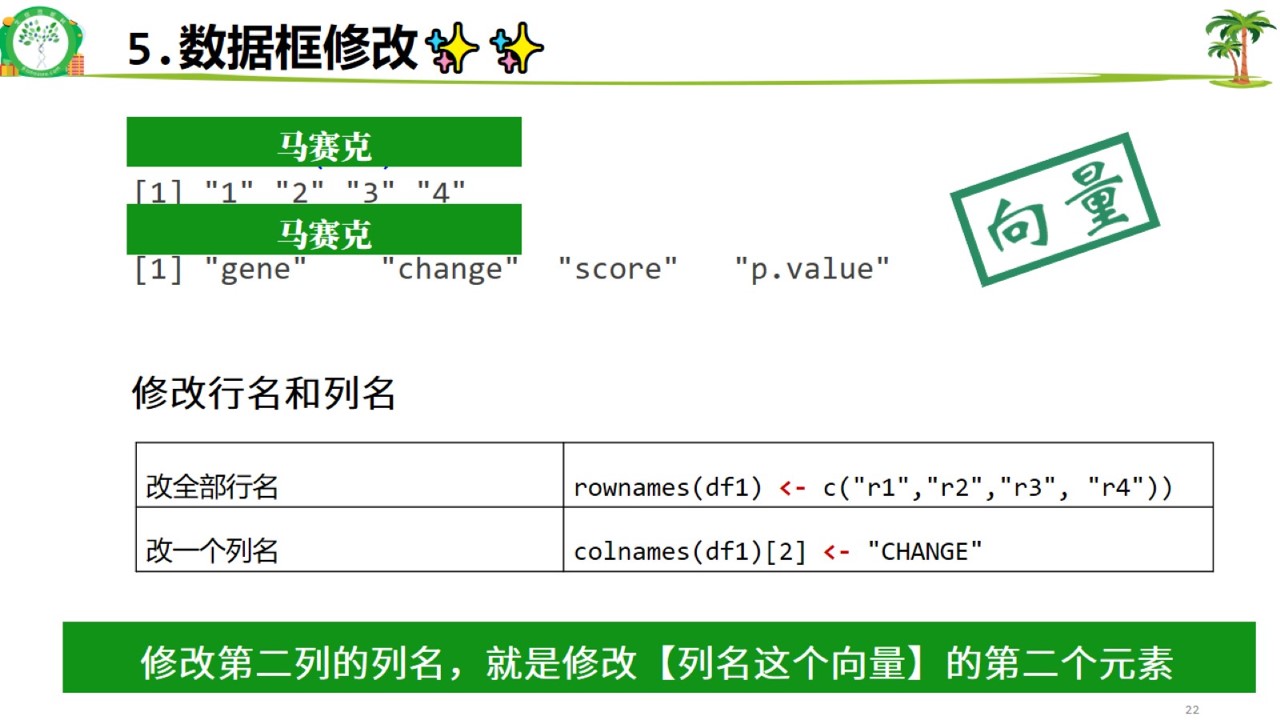
修改行列名.jpg
#改行名和列名
rownames(df1) <- c("r1","r2","r3","r4")
#只修改某一行/列的名
colnames(df1)[2] <- "CHANGE"6. 两个数据框的连接
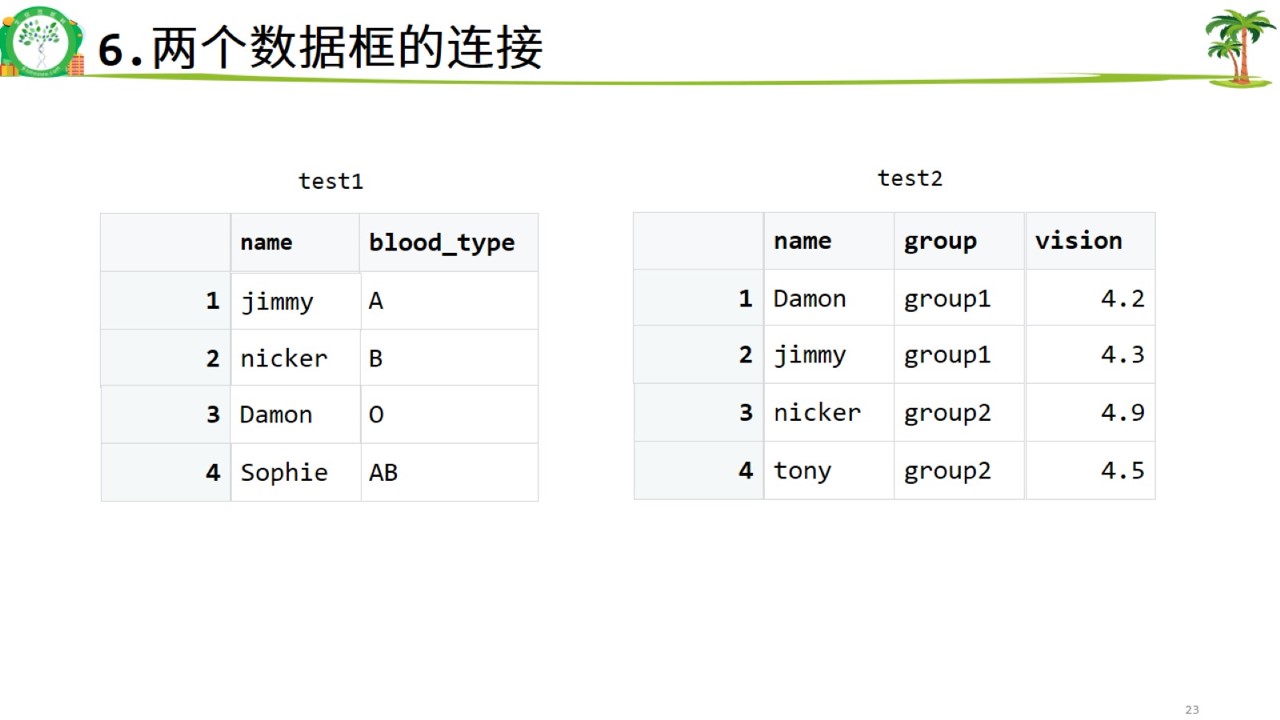
数据框连接.jpg
#两个数据框的连接
test1 <- data.frame(name = c('jimmy','nicker','Damon','Sophie'),
blood_type = c("A","B","O","AB"))
test1## name blood_type
## 1 jimmy A
## 2 nicker B
## 3 Damon O
## 4 Sophie ABtest2 <- data.frame(name = c('Damon','jimmy','nicker','tony'),
group = c("group1","group1","group2","group2"),
vision = c(4.2,4.3,4.9,4.5))
test2## name group vision
## 1 Damon group1 4.2
## 2 jimmy group1 4.3
## 3 nicker group2 4.9
## 4 tony group2 4.5test3 <- data.frame(NAME = c('Damon','jimmy','nicker','tony'),
weight = c(140,145,110,138))
test3## NAME weight
## 1 Damon 140
## 2 jimmy 145
## 3 nicker 110
## 4 tony 138merge(test1,test2,by="name")## name blood_type group vision
## 1 Damon O group1 4.2
## 2 jimmy A group1 4.3
## 3 nicker B group2 4.9merge(test1,test3,by.x = "name",by.y = "NAME")## name blood_type weight
## 1 Damon O 140
## 2 jimmy A 145
## 3 nicker B 1107.记忆卡片

记忆卡片3.jpg
8. 矩阵新建和取子集
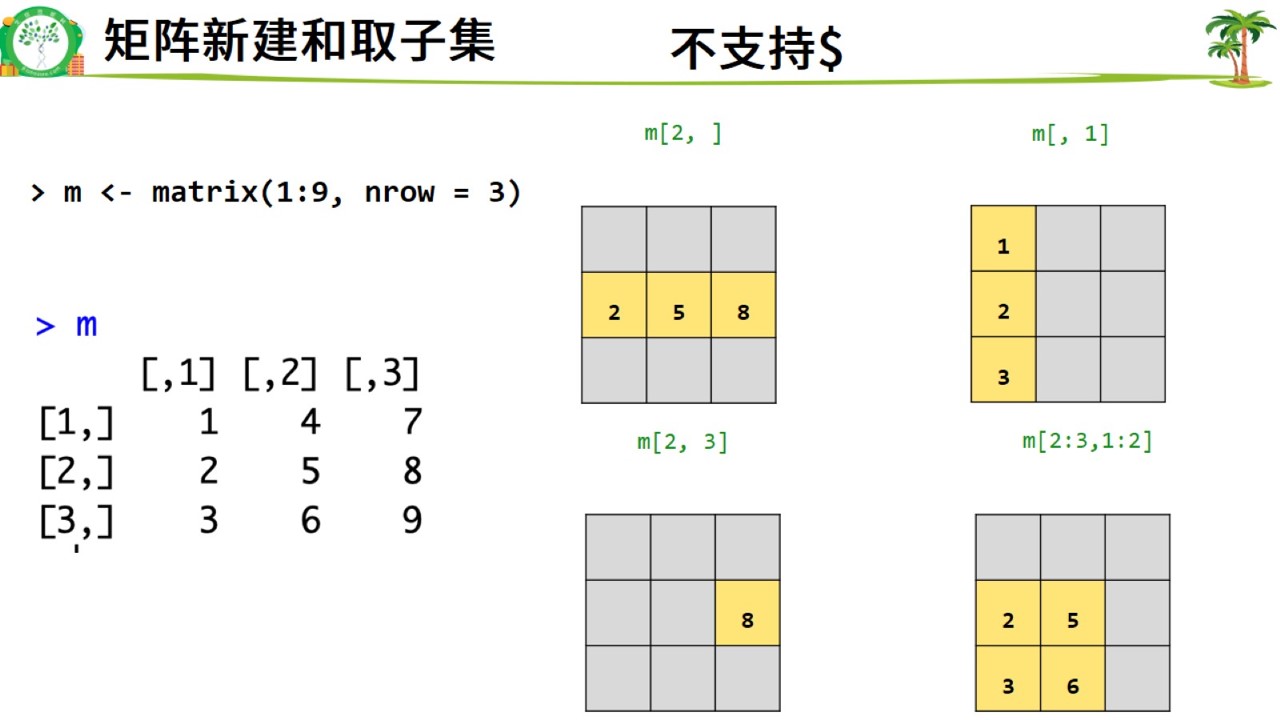
矩阵新建.jpg
m <- matrix(1:9, nrow = 3)
colnames(m) <- c("a","b","c") #加列名
m## a b c
## [1,] 1 4 7
## [2,] 2 5 8
## [3,] 3 6 9m[2,]## a b c
## 2 5 8m[,1]## [1] 1 2 3m[2,3]## c
## 8m[2:3,1:2]## a b
## [1,] 2 5
## [2,] 3 6m## a b c
## [1,] 1 4 7
## [2,] 2 5 8
## [3,] 3 6 99. 矩阵的转置和转换

转置和转换.jpg
t(m)## [,1] [,2] [,3]
## a 1 2 3
## b 4 5 6
## c 7 8 9class(m)## [1] "matrix" "array"m1 <- as.data.frame(m)
m1## a b c
## 1 1 4 7
## 2 2 5 8
## 3 3 6 9class(m1)## [1] "data.frame"10. 关于R语言的修改

R修.jpg
11. 列表的新建与取子集
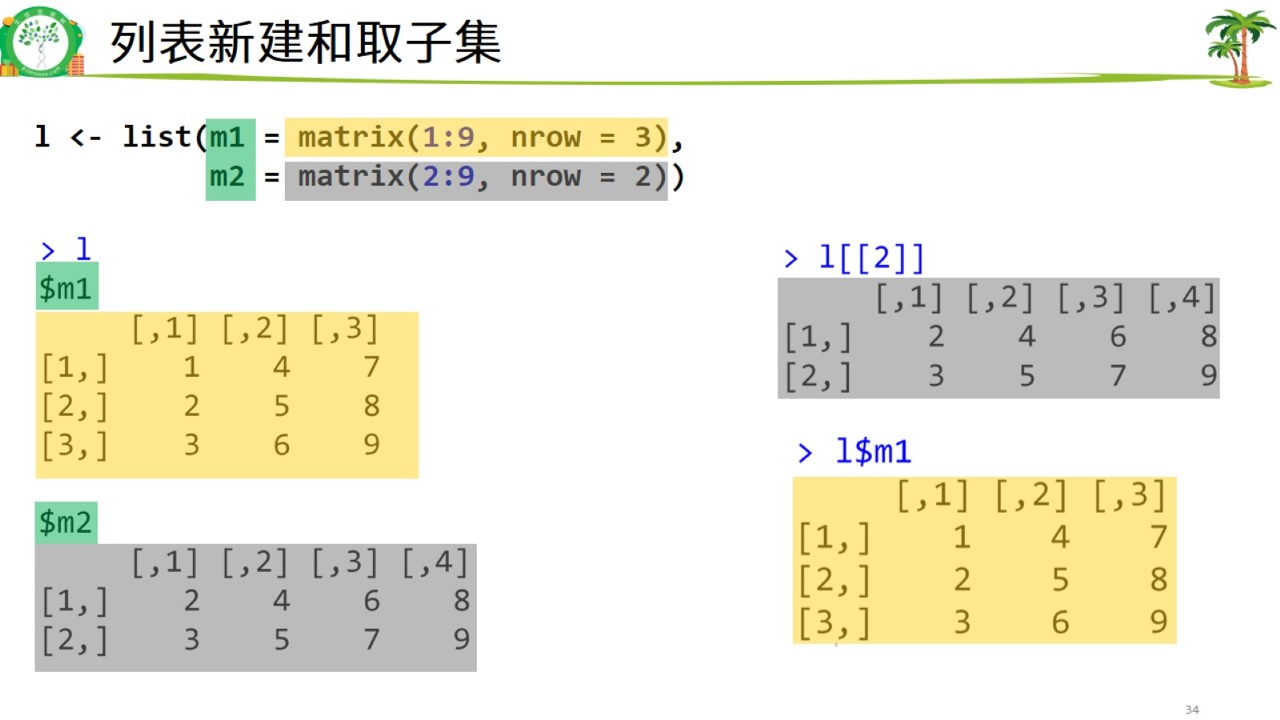
列表新建.jpg
#列表
l <- list(m1 = matrix(1:9, nrow = 3),
m2 = matrix(2:9, nrow = 2))
l## $m1
## [,1] [,2] [,3]
## [1,] 1 4 7
## [2,] 2 5 8
## [3,] 3 6 9
##
## $m2
## [,1] [,2] [,3] [,4]
## [1,] 2 4 6 8
## [2,] 3 5 7 9l[[2]]## [,1] [,2] [,3] [,4]
## [1,] 2 4 6 8
## [2,] 3 5 7 9l$m1## [,1] [,2] [,3]
## [1,] 1 4 7
## [2,] 2 5 8
## [3,] 3 6 9# 补充:元素的名字
scores = c(100,59,73,95,45)
names(scores) = c("jimmy","nicker","Damon","Sophie","tony")
scores## jimmy nicker Damon Sophie tony
## 100 59 73 95 45scores["jimmy"]## jimmy
## 100scores[c("jimmy","nicker")]## jimmy nicker
## 100 59names(scores)[scores>60]## [1] "jimmy" "Damon" "Sophie"12. 删除变量
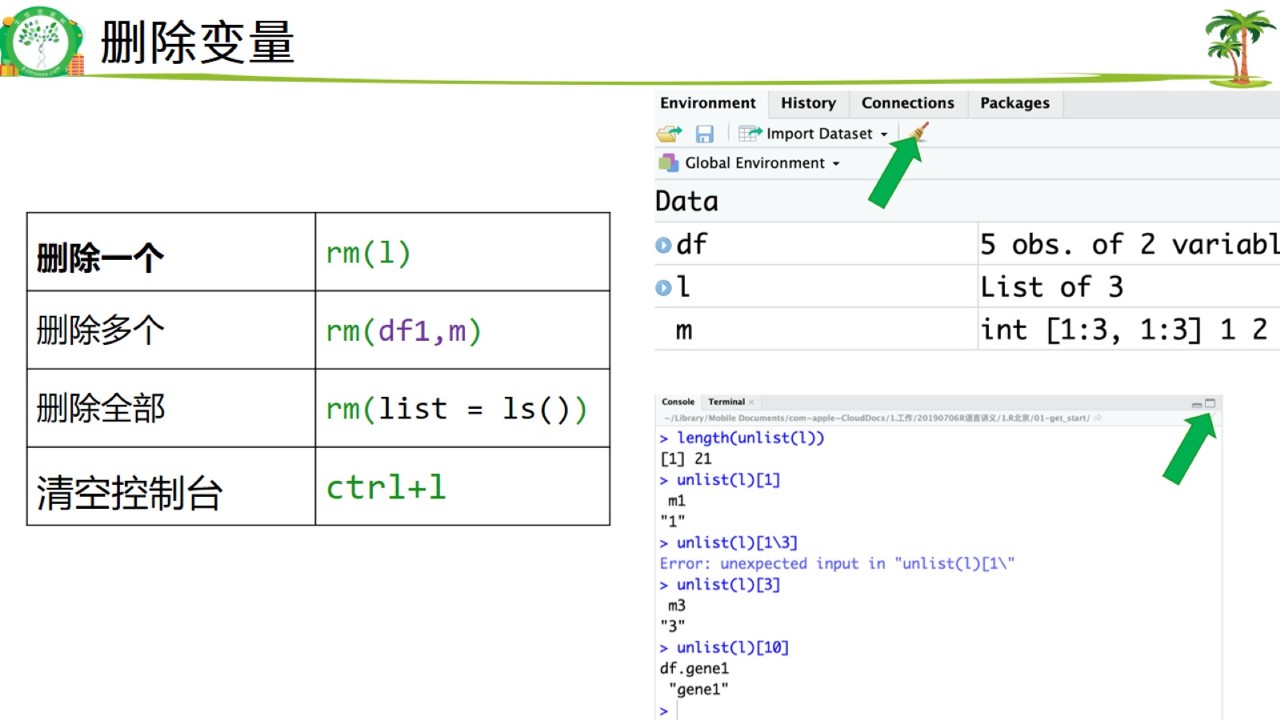
删除变量.jpg
13. 数据结构总结
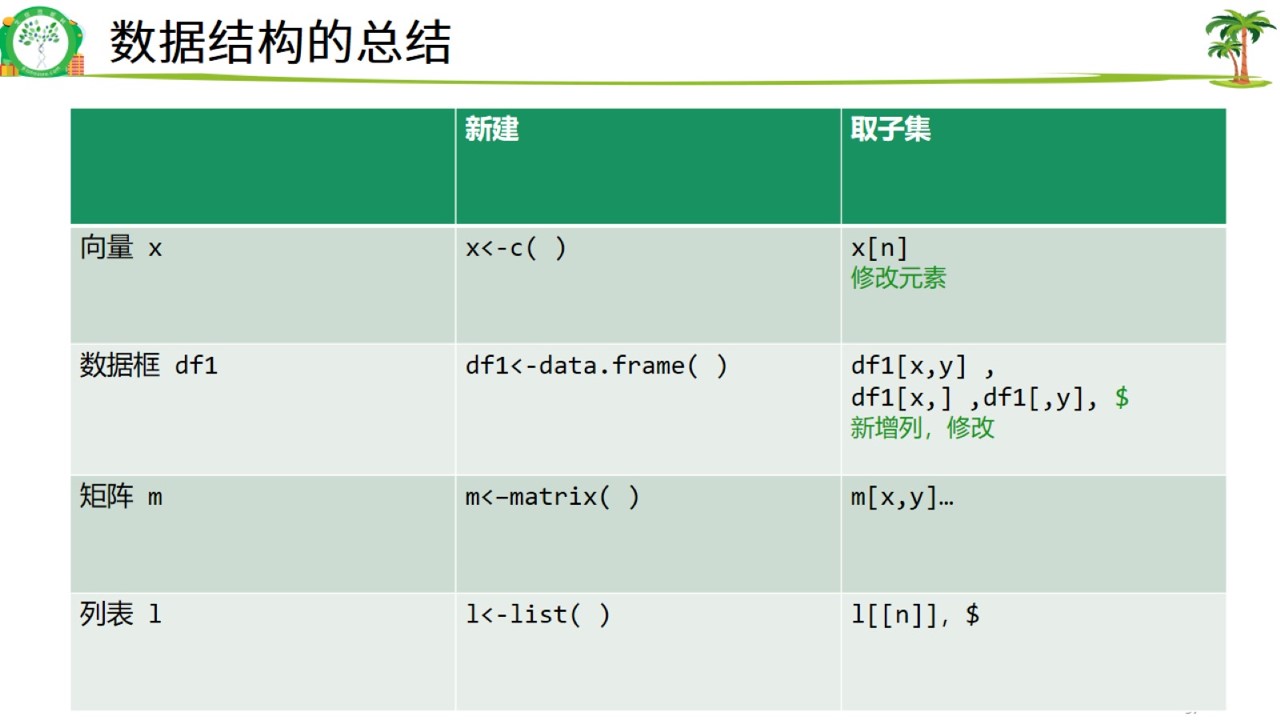
数据结构总结.jpg
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录

