单细胞学习小组003期 Day6
原创The content of today is marker gene and and cell annotation.
1. Loading data
this step is following the step yesterday.
I will use the data set of homework (GSESM7306055_sample2) for this step.
library(Seurat)
p1 = DimPlot(seu.obj, reduction = "umap", label = T)
p1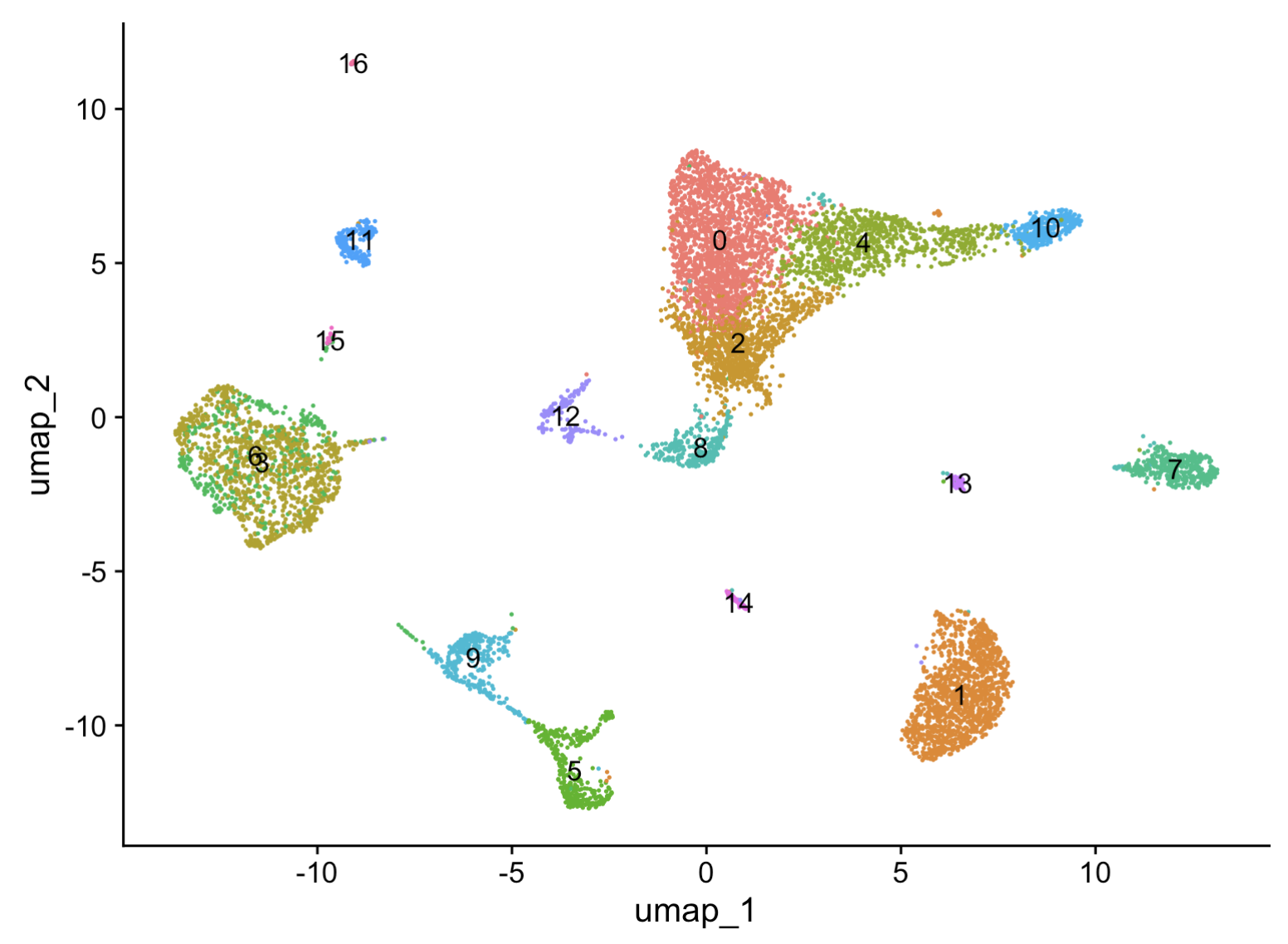
In this umap, cells were clustered into 16 clusters, which need to be annotated using marker genes.
2. Marker gene
Marker genes are the cell-specific genes, which are expressed high in corresponding cells but low in others. By marker genes, cell clusters can be annotated as specific cell types.
Theoretically, up- or down-regulated genes are all marker genes, but we prefer to make only.pos = TRUE, to focus on up-regulated genes as markers.
FindAllMarker is another speed-limiting step, which will be lagged by ammount of cells. It will calculate marker genes for all cell clusters, e.g., for cluster 0 through 0 vs 1 to 16 considering 1-16 as one group.
min.pct means that the gene is expressed in above 25% cells of the corresponding cluster.
library(dplyr)
f = "markers.Rdata"
if(!file.exists(f)){
markers <- FindAllMarkers(seu.obj, only.pos = TRUE,min.pct = 0.25) # calculate the marker genes for each cluster
save(markers,file = f)
}
load(f)2.1 How to install the R packages in github
Github is a private storage of developers. The R packages in Github can be downloaded and installed for use.
Two options to install:
install online
if(!require(devtools))install.packages("devtools")
if(!require(presto))devtools::install_github("immunogenomics/presto",upgrade = F,dependencies = T)or download first then install
if(!require(presto))devtools::install_github("presto-master.zip",upgrade = F,dependencies = T)2.2 Get top 2 log2|FC| marker genes for each cluster.
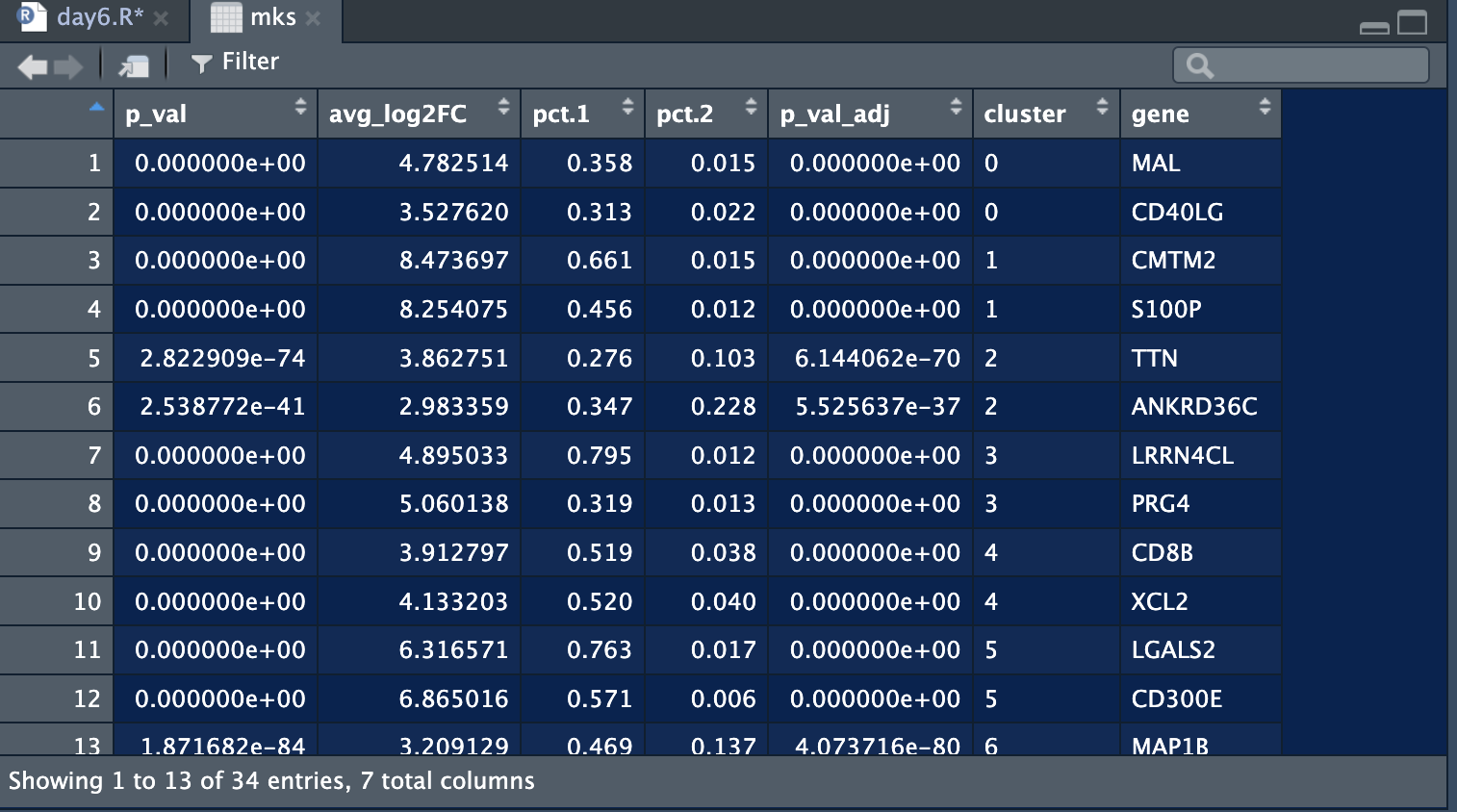
3. Five ways to visualise the marker genes
3.1 Heat map
library(ggplot2)
DoHeatmap(seu.obj, features = g) + NoLegend()+
scale_fill_gradientn(colors = c("#2fa1dd", "white", "#f87669"))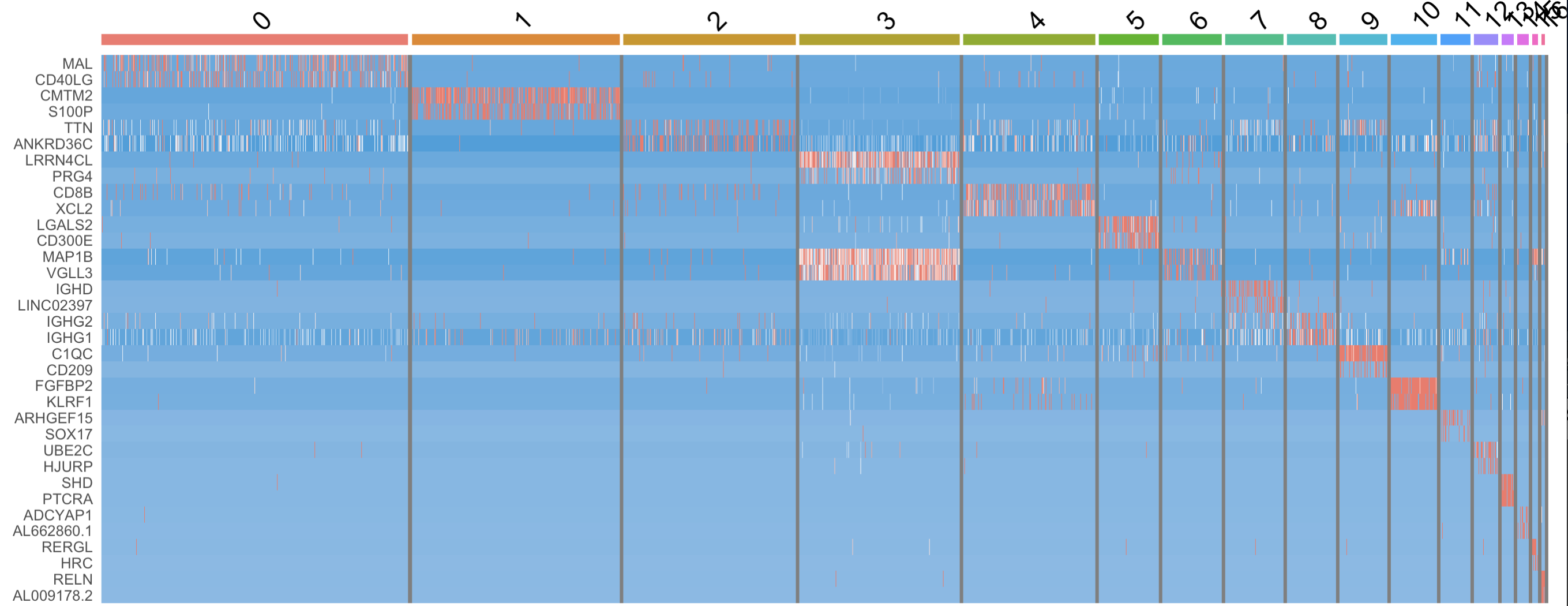
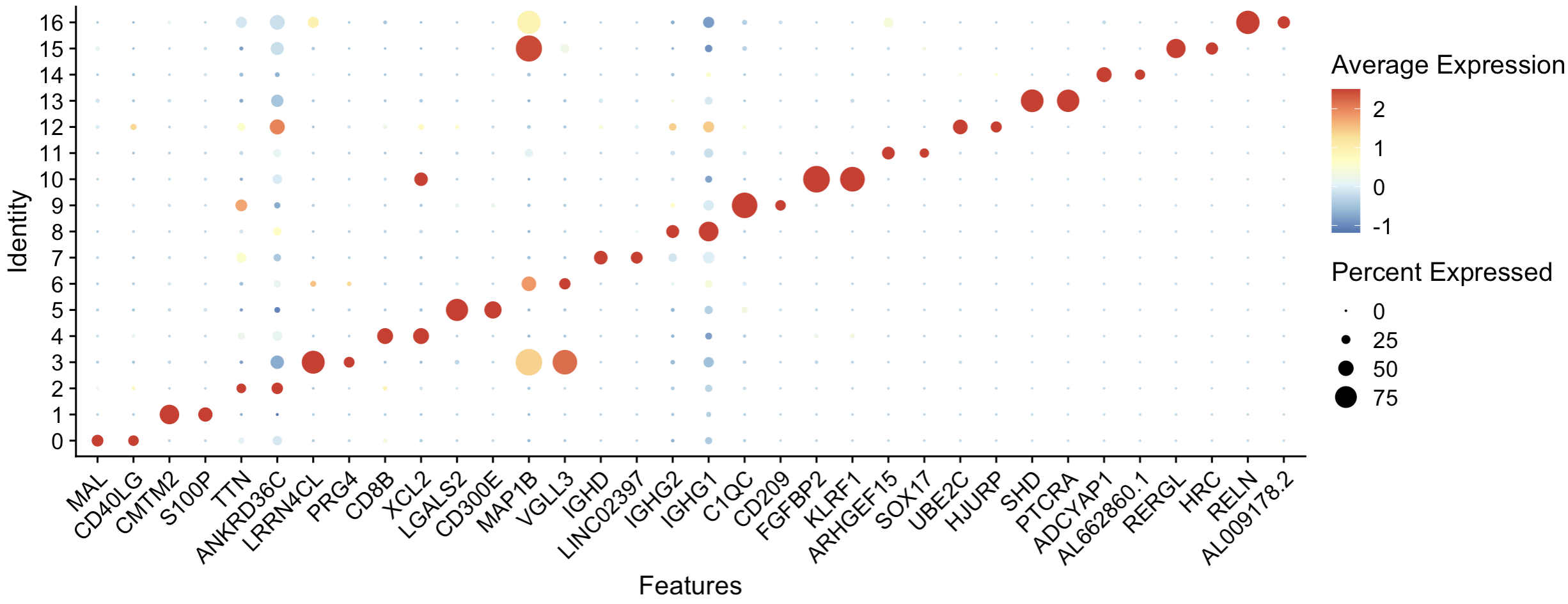
3.2 Bubble diagram
DotPlot(seu.obj, features = g,cols = "RdYlBu") +
RotatedAxis()Red presents up-regulated expression, the darker the higher. Size presents this gene's expression rate in the cells of this cluster.
3.3 Violin plot
VlnPlot(seu.obj, features = g[1:2])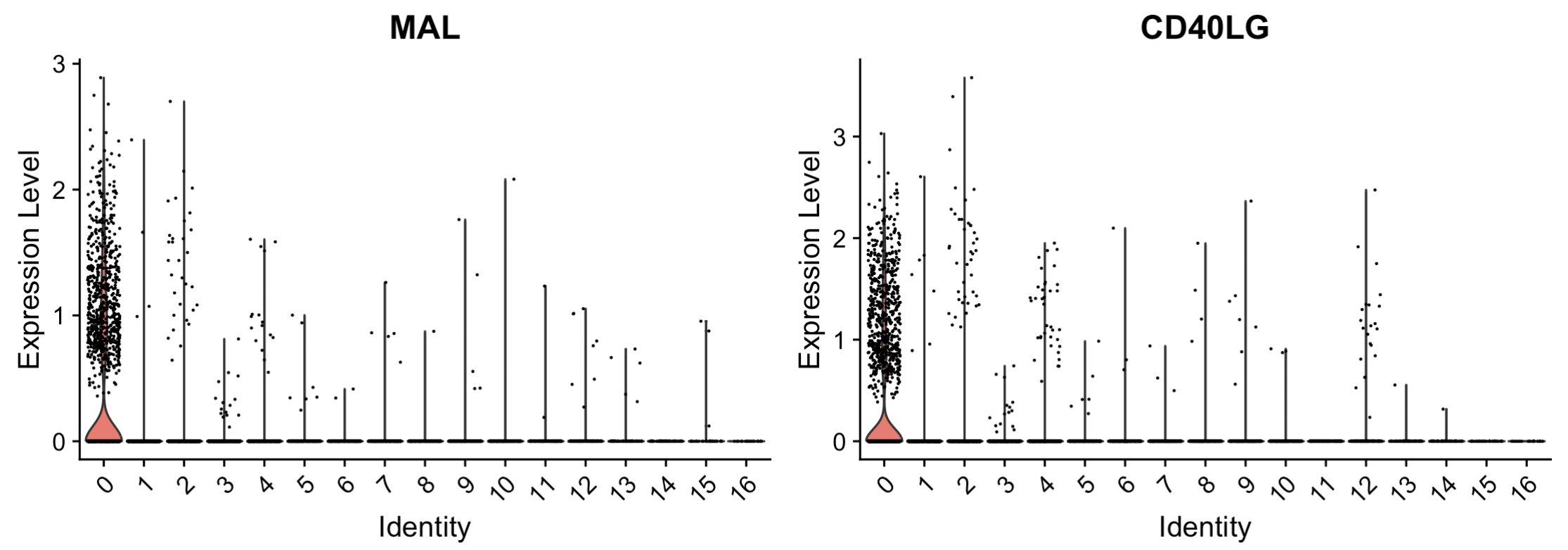
3.4 Feature plot
FeaturePlot(seu.obj, features = g[1:4])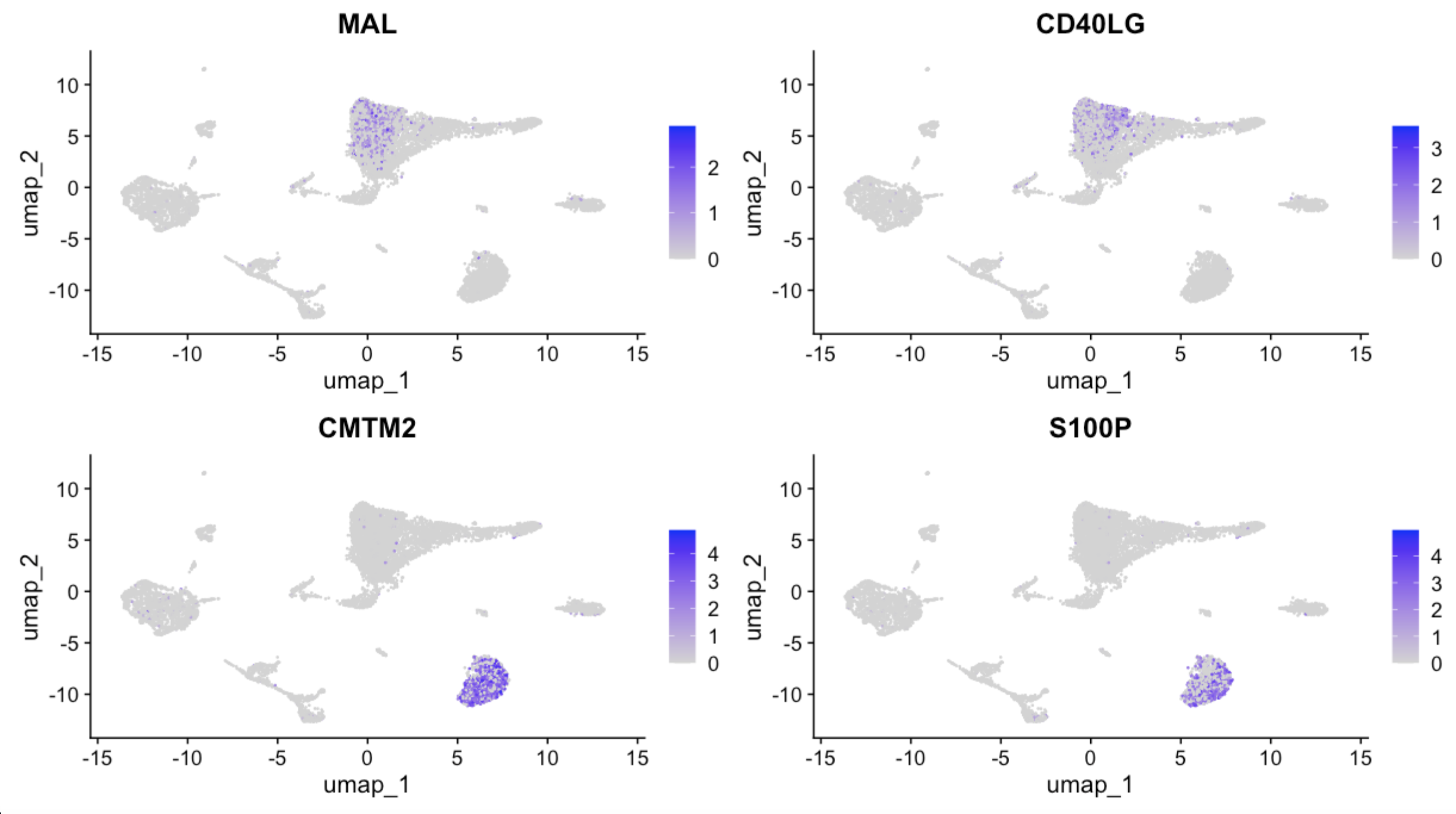
3.5 Peak map
RidgePlot(seu.obj, features = g[1:2])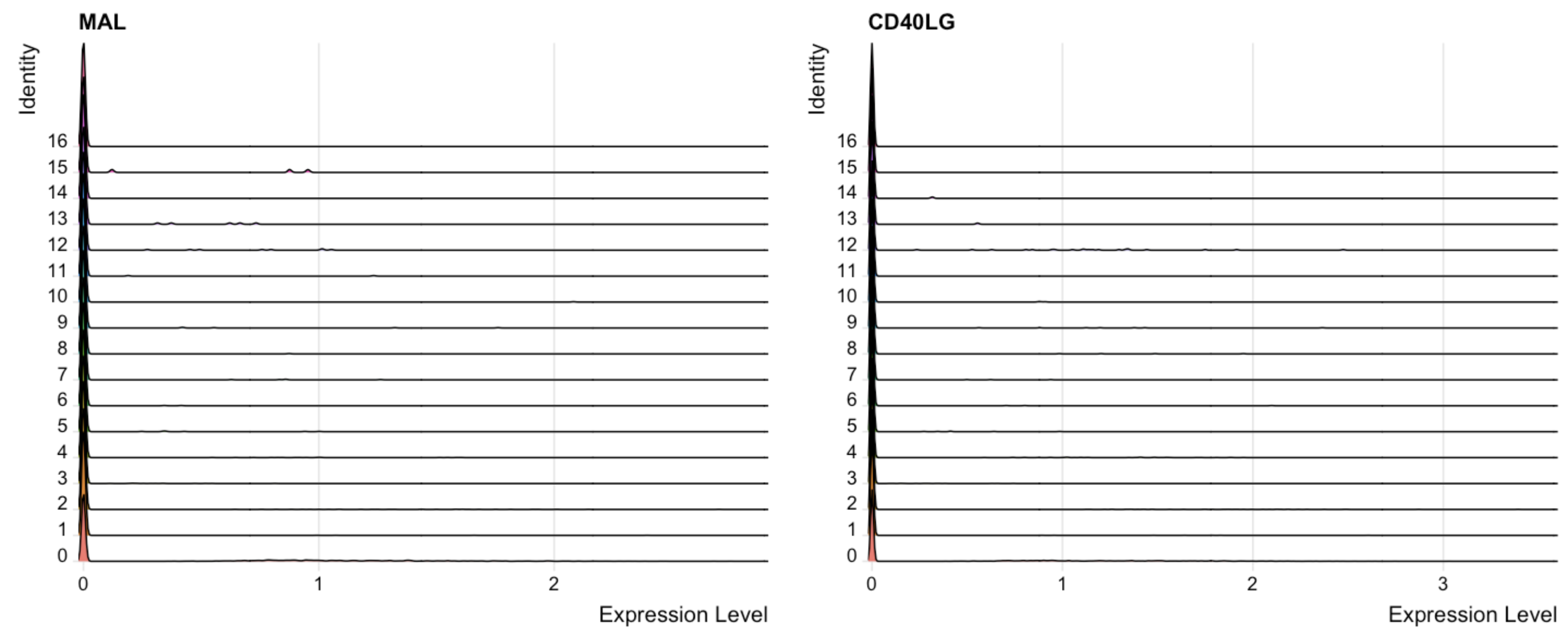
4. Annotation
manually or automatically
4.1 Manually, needs background knowledge
Search from literature or databases:
http://biocc.hrbmu.edu.cn/CellMarker
https://www.gsea-msigdb.org/gsea/msigdb/human/genesets.jsp?collection=C8
If use published data, check the related article for the marker genes used by the authors.
Nice tips from Huahua, a marker gene list, named my_markers.txt. This list should be storaged in the working directory.
I copied the list into a txt file and gave a same name as my_markers.txt.
Huahua used commas to seperate the columns instead of spaces, which can avoid the confusing space from some cell names.
a = read.table("my_markers.txt",sep = ",") # read my_markers
gt = split(a[,2],a[,1]) #split my_markers from long table into short table
DotPlot(seu.obj, features = gt,cols = "RdYlBu") +
RotatedAxis()
unique(a$V1) # list all cell types in my_markers.txt for later copy+paste
Make a anno.txt file
writeLines(paste0(0:16,","))Redifine the Seurat object by new idents
celltype = read.table("anno.txt",sep = ",")
celltype
levels(Idents(seu.obj))
levels(seu.obj)new.cluster.ids <- celltype$V2
names(new.cluster.ids) <- levels(seu.obj)
sce <- RenameIdents(seu.obj, new.cluster.ids)
save(sce,file = "sce.Rdata")
p2 <- DimPlot(sce, reduction = "umap",
label = TRUE, pt.size = 0.5) + NoLegend()
p1+p2
```R上传失败:Cannot read properties of undefined (reading 'url')
4.2 Automatic annotation
The most popolar package is SingleR, using reference data from celldex.
library(celldex)
library(SingleR)
ls("package:celldex")1 "BlueprintEncodeData"
2 "DatabaseImmuneCellExpressionData"
3 "defineTextQuery"
4 "fetchLatestVersion" # mouse
5 "fetchMetadata"
6 "fetchReference" # mouse
7 "HumanPrimaryCellAtlasData"
8 "ImmGenData"
9 "listReferences"
10 "listVersions"
11 "MonacoImmuneData"
12 "MouseRNAseqData"
13 "NovershternHematopoieticData"
14 "saveReference"
15 "searchReferences"
16 "surveyReferences"
Except for 4 and 6, others are for human.
f = "ref_BlueprintEncode.RData"
if(!file.exists(f)){
ref <- celldex::BlueprintEncodeData()
save(ref,file = f)
}
ref <- get(load(f))library(BiocParallel)
scRNA = seu.obj
test = scRNA@assays$RNA$data
pred.scRNA <- SingleR(test = test,
ref = ref,
labels = ref$label.main,
clusters = scRNA@active.ident)
pred.scRNA$pruned.labels
## [1] "B-cells" "CD8+ T-cells" "CD4+ T-cells"
## [4] "CD4+ T-cells" "CD4+ T-cells" "B-cells"
## [7] "Monocytes" "Endothelial cells" "Fibroblasts"
## [10] "NK cells" "Endothelial cells" NA
new.cluster.ids <- pred.scRNA$pruned.labels
names(new.cluster.ids) <- levels(scRNA)
scRNA <- RenameIdents(scRNA,new.cluster.ids)
p3 <- DimPlot(scRNA, reduction = "umap",label = T,pt.size = 0.5) + NoLegend()
p2+p3The last picture p2+p3 failed to upload many times. Quit at last.
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
