跟着Nature学空转分析(一):10x visium多样本基础分析的再次回顾---解析生孩子的困境
跟着Nature学空转分析(一):10x visium多样本基础分析的再次回顾---解析生孩子的困境

KS科研分享与服务-TS的美梦
发布于 2025-12-20 17:28:21
发布于 2025-12-20 17:28:21
# ---------------------------------------------------------------
# ----- -----
# ----- 10X Visium 空转分析教程【seurat】 -----
# ----- 《KS科研分享与服务》公众号出品 -----
# ----- -----
# ----- -----
# ----- 第一节:基础分析 -----
# ----- -----
# ----------------------------------------------------------------
#!!!!此分析基于seurat v5!!!!上游数据分析参考:
空转上游你可以不做,但不能不会呀!详细演示Visium HD上游公共数据库分析及图像对齐矫正
对于visium的分析之前出过,虽然现在visium HD都很普遍了,但是对于经费有限的课题组,visium还是主流,且随着公共数据库数据集的增大,后期还是有很大的挖掘潜力,所以有必要做一次完成的教程介绍。可能您已经接触过空转visium分析,或者看到过很多教程,本次我们的教程将采取最新版本的软件进行分析。我们的风格还是出能让人看懂的教程,从简单到复杂,不给小白堵死们!
1、shell-download data
数据集是发表在Zhang, Y., Pan, J., Liu, Z. et al. A spatial transcriptomics dataset of the endometrium from repeated implantation failure patients. Sci Data 12, 1711 (2025). https://doi.org/10.1038/s41597-025-05995-6的数据,这篇文章的空转数据没有做太多深入的分析,一共有8个样本,这里挑选了4个演示。样本有两个组,一组是正常人的子宫内膜样本,另外一组是RIF即反复植入失败的样本。目前环境下,由于各种因素,其实生殖是一个很重大且很难的事情,恰好博主以前的专业涉及过生殖,所以选取相关样本演示:
#=================================================================================
# nohup wget https://ftp.ncbi.nlm.nih.gov/geo/series/GSE287nnn/GSE287278/suppl/GSE287278_RAW.tar &
# cd data_analysis/Spatial_analysis/visium/
# tar -xvf GSE287278_RAW.tar
# rm GSE287278_RAW.tar
# for f in GSM*.tar.gz; do
# tar -xzf "$f"
# done
#rm GSM*.tar.gz
#ls -lh
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 CTR1_processed_data
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 CTR2_processed_data
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 CTR3_processed_data
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 CTR4_processed_data
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 RIF1_processed_data
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 RIF2_processed_data
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 RIF3_processed_data
# drwxr-xr-x 4 ks_ts ks_ts 4.0K Dec 19 2024 RIF4_processed_data2、读取数据
#加载数据用的是Load10X_Spatial,?Load10X_Spatial可以查看它的帮助函数
#这里加载的其实是标准的10X空转下级数据,Load10X_Spatial示例也是最标准的读取方式
#如果你的数据是10X spaceranger得到的,下游文件在out文件夹,很容易进行读取
#但是如果不跑上游,也没有标准的文件,例如使用公共数据库数据,可能就需要各种形式处理了
# Load10X_Spatial(
# data.dir, #h5文件也就是表达矩阵文件夹,image文件在其下级文件夹spatial
# filename = "filtered_feature_bc_matrix.h5",#feature barcode matrix文件名
# assay = "Spatial",
# slice = "slice1",
# filter.matrix = TRUE,只保留确定在组织上的spots
# to.upper = FALSE,
# image = NULL,
# ...
# )
#标准文件读取
#路径应该包含filtered_feature_bc_matrix.h5文件以及spatial文件夹
# sp <- Load10X_Spatial(data.dir = './sta_files/',
# filename = "filtered_feature_bc_matrix.h5",
# assay = "Spatial")setwd("~/data_analysis/Spatial_analysis/visium/")
#加载包
library(Seurat)
library(ggplot2)
library(patchwork)
library(dplyr)
library(SeuratWrappers)因为没有跑上游,我们演示的数据作者提供的是spatial文件夹,但是表达矩阵是10x标准的3个文件,所以数据读取需要分别进行。首先使用Read10X读取表达矩阵并创建seurat obj,与scRNA是一样的,然后使用Read10X_Image读取image信息,此函数专门用于读取visium图片信息,visium1和visium HD都适用!原文提供了8个sample,演示在control和RIF组各选了2个sample进行分析示例。
#control3 sample
CTR3 <- CreateSeuratObject(counts = Read10X("./CTR3_processed_data/filtered_feature_bc_matrix/"), assay = "Spatial",project='CTR3')#读取exp matrix,并创建seurat obj
CTR3_img <- Read10X_Image(image.dir = file.path("./CTR3_processed_data/spatial/"), filter.matrix = TRUE)#Load a 10X Genomics Visium Image
CTR3_img <- CTR3_img[Cells(x = CTR3)]#保留图像中与表达矩阵一致的spot
DefaultAssay(CTR3) <- "Spatial"
CTR3[["CTR3"]] <- CTR3_img
#control4 sample
CTR4 <- CreateSeuratObject(counts = Read10X("./CTR4_processed_data/filtered_feature_bc_matrix/"), assay = "Spatial",project='CTR4')#读取exp matrix,并创建seurat obj
CTR4_img <- Read10X_Image(image.dir = file.path("./CTR4_processed_data/spatial/"), filter.matrix = TRUE)#Load a 10X Genomics Visium Image
CTR4_img <- CTR4_img[Cells(x = CTR4)]#保留图像中与表达矩阵一致的spot
DefaultAssay(CTR4) <- "Spatial"
CTR4[["CTR4"]] <- CTR4_img
#RIF3 sample
RIF3 <- CreateSeuratObject(counts = Read10X("./RIF3_processed_data/filtered_feature_bc_matrix/"), assay = "Spatial",project='RIF3')#读取exp matrix,并创建seurat obj
RIF3_img <- Read10X_Image(image.dir = file.path("./RIF3_processed_data/spatial/"), filter.matrix = TRUE)#Load a 10X Genomics Visium Image
RIF3_img <- RIF3_img[Cells(x = RIF3)]#保留图像中与表达矩阵一致的spot
DefaultAssay(RIF3) <- "Spatial"
RIF3[["RIF3"]] <- RIF3_img
#RIF4 sample
RIF4 <- CreateSeuratObject(counts = Read10X("./RIF4_processed_data/filtered_feature_bc_matrix/"), assay = "Spatial",project='RIF4')#读取exp matrix,并创建seurat obj
RIF4_img <- Read10X_Image(image.dir = file.path("./RIF4_processed_data/spatial/"), filter.matrix = TRUE)#Load a 10X Genomics Visium Image
RIF4_img <- RIF4_img[Cells(x = RIF4)]#保留图像中与表达矩阵一致的spot
DefaultAssay(RIF4) <- "Spatial"
RIF4[["RIF4"]] <- RIF4_img
#remove
rm(CTR3_img,CTR4_img,RIF3_img,RIF4_img)3、可视化基本信息
#可视化spots ncount/nfeatures,以检查数据以及构建的object无误
SpatialFeaturePlot(CTR3, features = "nCount_Spatial")+ theme(legend.position = "right")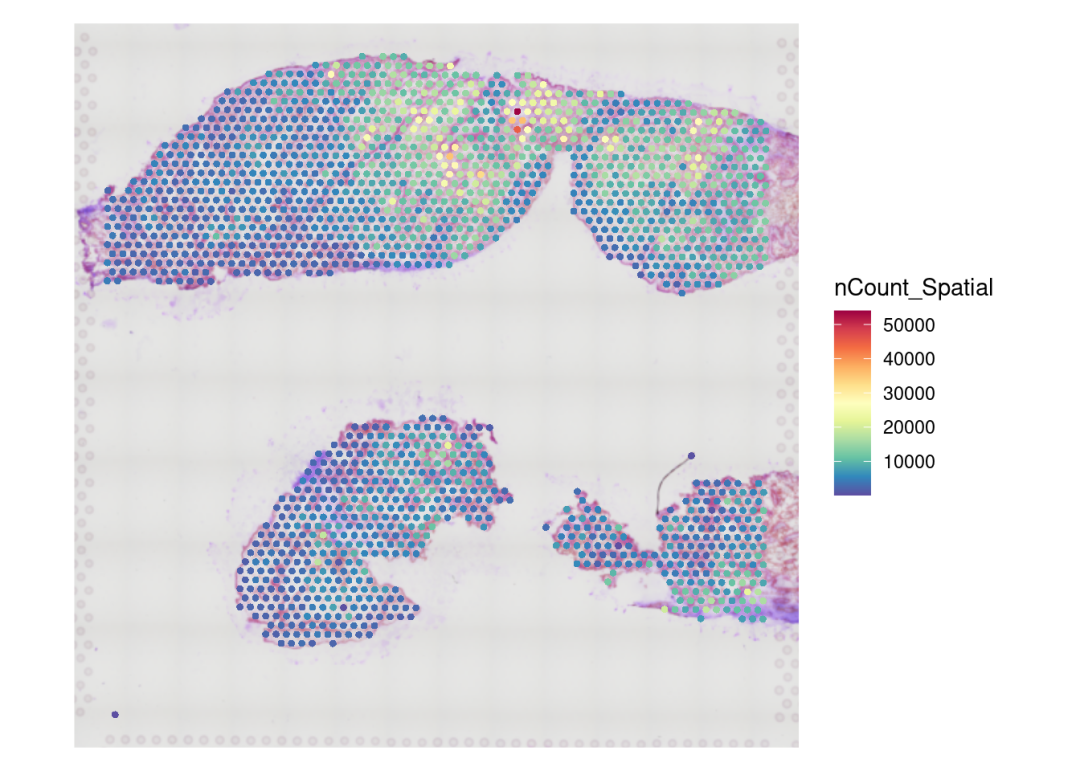
SpatialFeaturePlot(RIF3, features = "nCount_Spatial") +theme(legend.position = "right")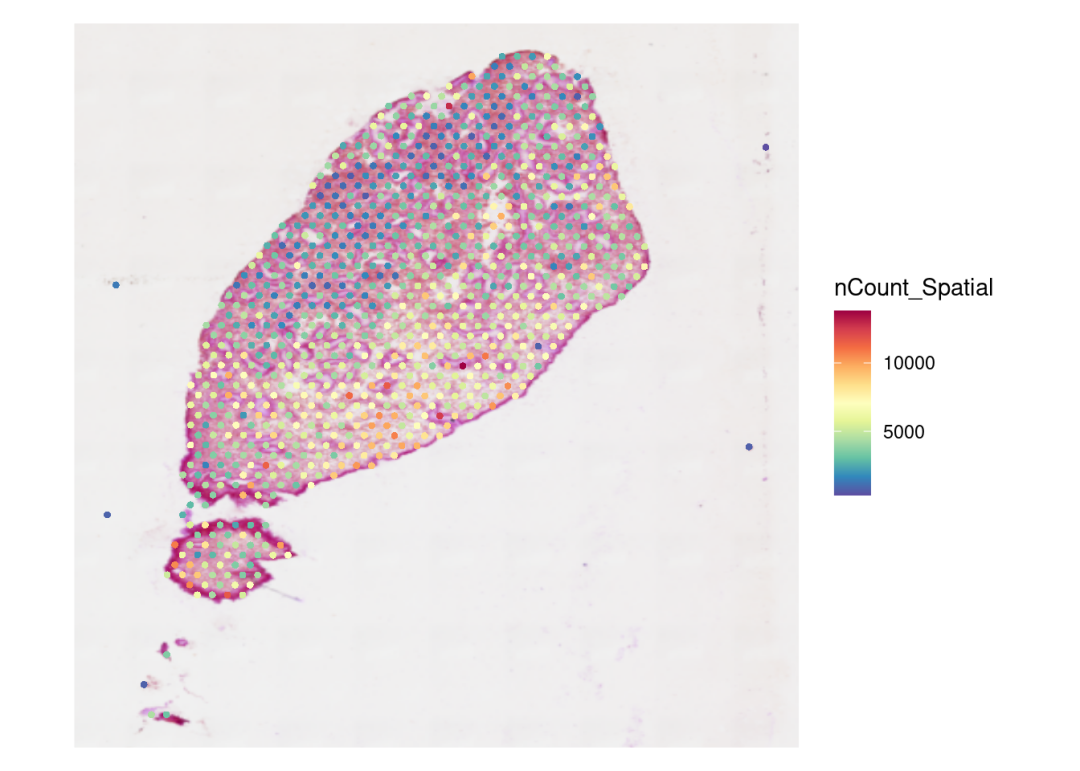
VlnPlot(CTR3, features = "nCount_Spatial", pt.size = 0.1)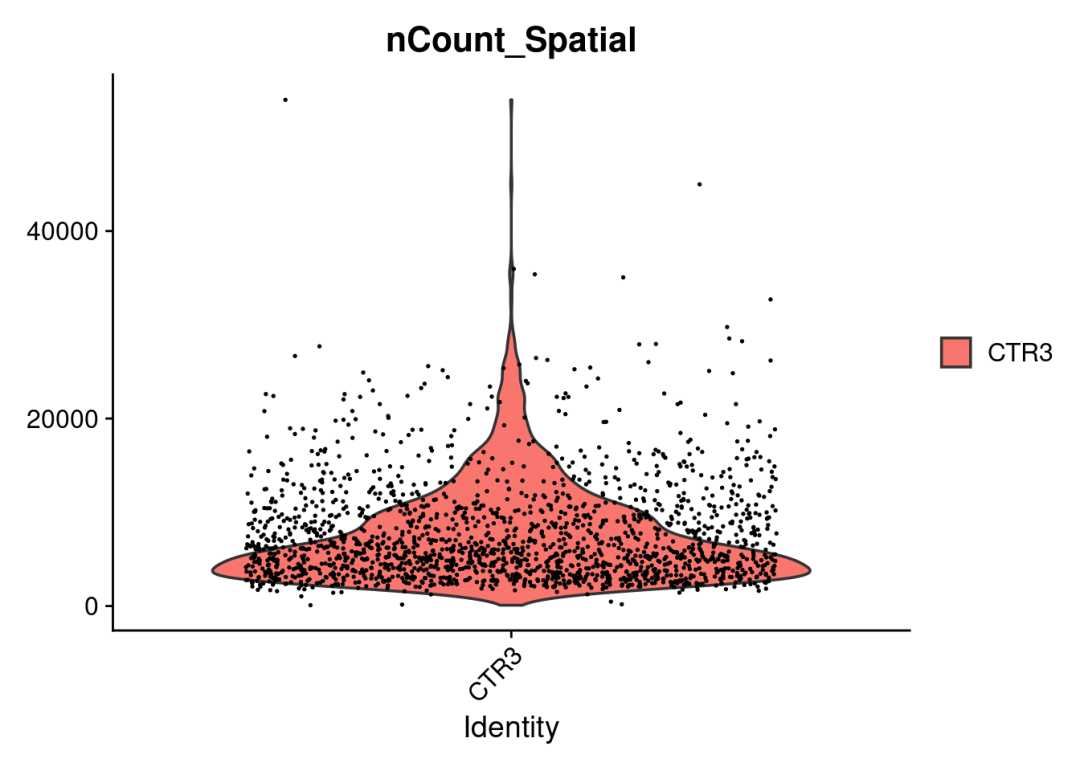
VlnPlot(RIF3, features = "nCount_Spatial", pt.size = 0.1)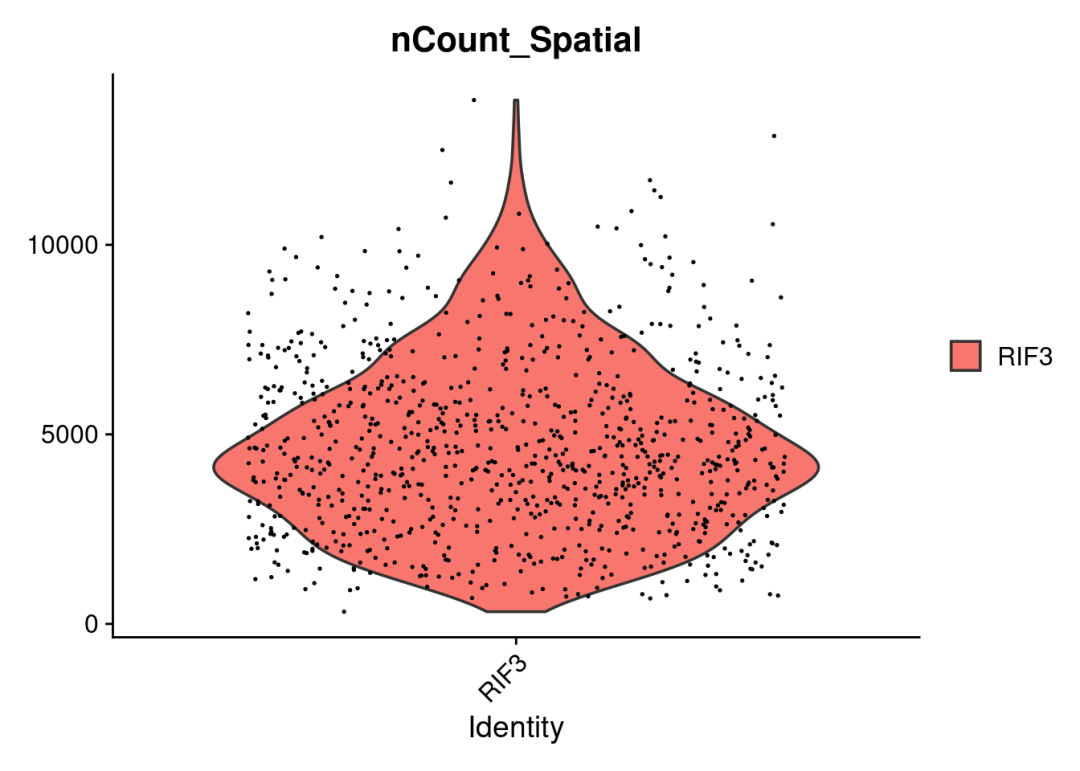
4、质控及标准化
#add metadata
#添加sample分组或者其他信息
CTR3$group <- 'CTR'
CTR3$group <- 'CTR'
RIF3$group <- 'RIF'
RIF4$group <- 'RIF'接下来就是数据质控以及标准化流程,这些流程与标准的scRNA流程非常类似。关于指控指标,也就是scRNA中提到的那些点,例如线体力基因比例、ncount范围等等(当然在visium中是针对spot的)。原文描述的质控指标:Spots with gene count below 500 or mitochondrial gene percentage exceeding 20% were excluded for each slice。合并不同的sample为一个list,便于处理:
obj.list <- list(CTR3,CTR4,RIF3,RIF4)
for (i in 1:length(obj.list)) {
obj.list[[i]][["percent.mt"]] <- PercentageFeatureSet(obj.list[[i]], pattern = "^MT-")#计算spot线粒体基因比例
obj.list[[i]] <- subset(obj.list[[i]], subset = nCount_Spatial > 500 & percent.mt < 20)
}接下来需要对数据进行归一化处理,以校正不同测序深度在各个数据点之间带来的差异。在空间转录组数据中,由于不同位置(spot)之间的细胞密度可能存在差异,每个spot的分子计数(molecular counts/spot)往往具有较大的变异性。所以要求使用有效的归一化方法来减少技术噪音并提高生物信号的可比性。归一化的方法上,建议使用SCTransform,效果更好,能够在校正技术噪音的同时,更好地保留真实的生物学变异。seurat官网还有对比(https://satijalab.org/seurat/articles/spatial_vignette)
# perform SCTransform normalization
for (i in 1:length(obj.list)) {
obj.list[[i]] <- SCTransform(obj.list[[i]], assay = 'Spatial'
#vars.to.regress = "percent.mt #指定需要回归掉的技术噪音变量
)
}5、数据整合 & 降维聚类
空转样本单独分析还是整合分析?
???关于多样本空转数据分析需不需要整合分析的看法??? 首先说结论然后再解释(这里纯属个人看法思考及参考了网上部分意见):空转样本整合分析非必需,但是如果多样本还是有必要!
空转数据如果你有两个及以上的样本,没有scRNA那种需要进行条件对比的数据,是没有整合去批次的需求的,完全可以单个sample分开进行分析。同时,空转数据每个切片都是不一样的,所以整合也没有必要,更不能说通过整合去去批次。之所以进行“整合”一说,对于空转指示对spot表达矩阵的整合,方便进行降维聚类、横向比较以及注释,如果不进行整合,那么merge的多个切片spot表达矩阵降维的UMAP cluster,每个sampl都是独立的,不利于分析。整合只是对spot表达矩阵的整合,用于后续的降维,并没有对image整合,就目前来说那是不可能的也是不可取的。
这里演示数据有4个sample,我们采取SCT+seurat的整合方式:
# Merge all samples
# Create RNA assay for all spatial objects
for (i in 1:length(obj.list)) {
obj.list[[i]][["RNA"]] <- obj.list[[i]][["Spatial"]]
}anchor_features <- SelectIntegrationFeatures(object.list = obj.list, nfeatures = 3000)
obj.list <- PrepSCTIntegration(object.list = obj.list, anchor.features = anchor_features)
rsc.anchors <- FindIntegrationAnchors(object.list = obj.list, normalization.method = "SCT",anchor.features = anchor_features) # Takes time!## Warning in CheckDuplicateCellNames(object.list = object.list): Some cell names
## are duplicated across objects provided. Renaming to enforce unique cell names.## Finding all pairwise anchors## Running CCA## Merging objects## Warning: Adding image data that isn't associated with any assays## Warning: Adding image data that isn't associated with any assays## Warning: Key 'slice1_' taken, using 'ctr4_' instead## Finding neighborhoods## Finding anchors## Found 6525 anchors## Filtering anchors## Retained 3041 anchors## Running CCA## Merging objects## Warning: Adding image data that isn't associated with any assays## Warning: Adding image data that isn't associated with any assays## Warning: Key 'slice1_' taken, using 'rif3_' instead## Finding neighborhoods## Finding anchors## Found 3985 anchors## Filtering anchors## Retained 2596 anchors## Running CCA## Merging objects## Warning: Adding image data that isn't associated with any assays## Warning: Adding image data that isn't associated with any assays## Warning: Key 'slice1_' taken, using 'rif3_' instead## Finding neighborhoods## Finding anchors## Found 4029 anchors## Filtering anchors## Retained 2574 anchors## Running CCA## Merging objects## Warning: Adding image data that isn't associated with any assays## Warning: Adding image data that isn't associated with any assays## Warning: Key 'slice1_' taken, using 'rif4_' instead## Finding neighborhoods## Finding anchors## Found 4811 anchors## Filtering anchors## Retained 2700 anchors## Running CCA## Merging objects## Warning: Adding image data that isn't associated with any assays## Warning: Adding image data that isn't associated with any assays## Warning: Key 'slice1_' taken, using 'rif4_' instead## Finding neighborhoods## Finding anchors## Found 4952 anchors## Filtering anchors## Retained 2907 anchors## Running CCA## Merging objects## Warning: Adding image data that isn't associated with any assays## Warning: Adding image data that isn't associated with any assays## Warning: Key 'slice1_' taken, using 'rif4_' instead## Finding neighborhoods## Finding anchors## Found 3520 anchors## Filtering anchors## Retained 2196 anchorsmerged <- IntegrateData(anchorset = rsc.anchors, normalization.method = "SCT", k.weight = 40)merged[["RNA"]] <- JoinLayers(merged[["RNA"]])降维聚类步骤与scRNA无异
merged <- RunPCA(merged, npcs = 30, verbose = FALSE)
merged <- RunUMAP(merged, reduction = "pca", dims = 1:30)## Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
## To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
## This message will be shown once per session## 18:54:53 UMAP embedding parameters a = 0.9922 b = 1.112## 18:54:53 Read 5883 rows and found 30 numeric columns## 18:54:53 Using Annoy for neighbor search, n_neighbors = 30## 18:54:53 Building Annoy index with metric = cosine, n_trees = 50## 0% 10 20 30 40 50 60 70 80 90 100%## [----|----|----|----|----|----|----|----|----|----|## **************************************************|
## 18:54:55 Writing NN index file to temp file /tmp/RtmptDpooK/file81ac7575aa159
## 18:54:55 Searching Annoy index using 1 thread, search_k = 3000
## 18:54:57 Annoy recall = 100%
## 18:54:59 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
## 18:55:01 Initializing from normalized Laplacian + noise (using RSpectra)
## 18:55:01 Commencing optimization for 500 epochs, with 289038 positive edges
## 18:55:01 Using rng type: pcg
## 18:55:12 Optimization finishedmerged <- FindNeighbors(merged, reduction = "pca", dims = 1:30)
merged <- FindClusters(merged, resolution = 0.6)6、一些基本结果可视化
查看整合的降维UMAP:
color_pal<-c("#11A579","#F2B701","#66C5CC","#80BA5A","#F6CF71","#7F3C8D","#CF1C90","#3969AC","#f97b72","#E73F74","#4b4b8f","#ACA4E2","#31C53F","#B95FBB","#D4D915","#28CECA")
DimPlot(merged, reduction = "umap", label = F, pt.size = 0.5,cols = color_pal)+
DimPlot(merged, reduction = "umap", group.by = 'orig.ident',label = F, pt.size = 0.5)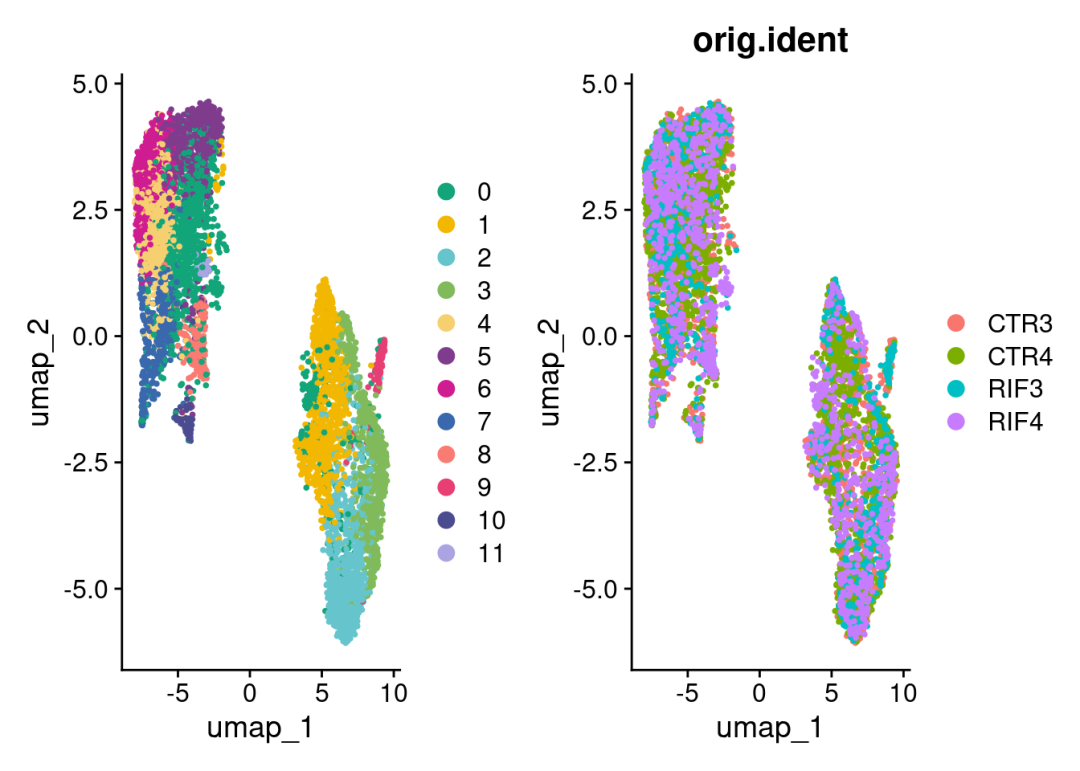
可视化slice:
SpatialDimPlot(merged, stroke=0.1,ncol = 2)& #如果spots不需要边框,设置stroke=NA
scale_fill_manual(values = color_pal) 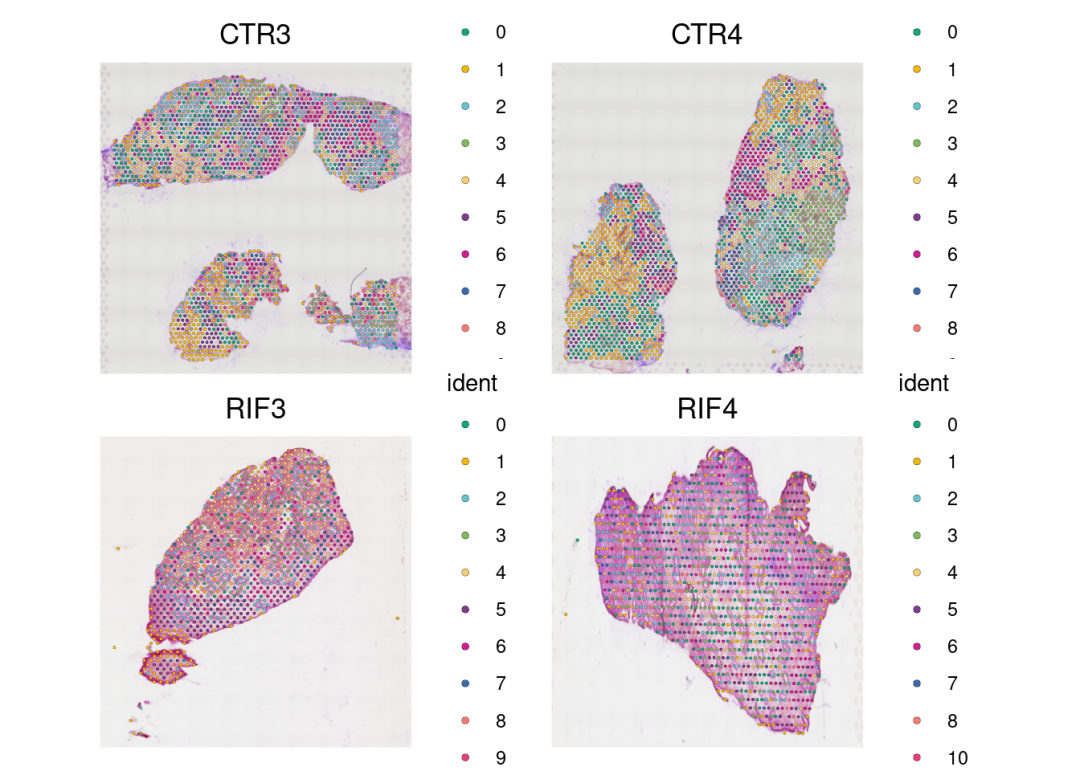
基因表达的空间分布:
DefaultAssay(merged) <- 'SCT'
SpatialFeaturePlot(merged, 'PAEP', ncol = 2, min.cutoff = 0)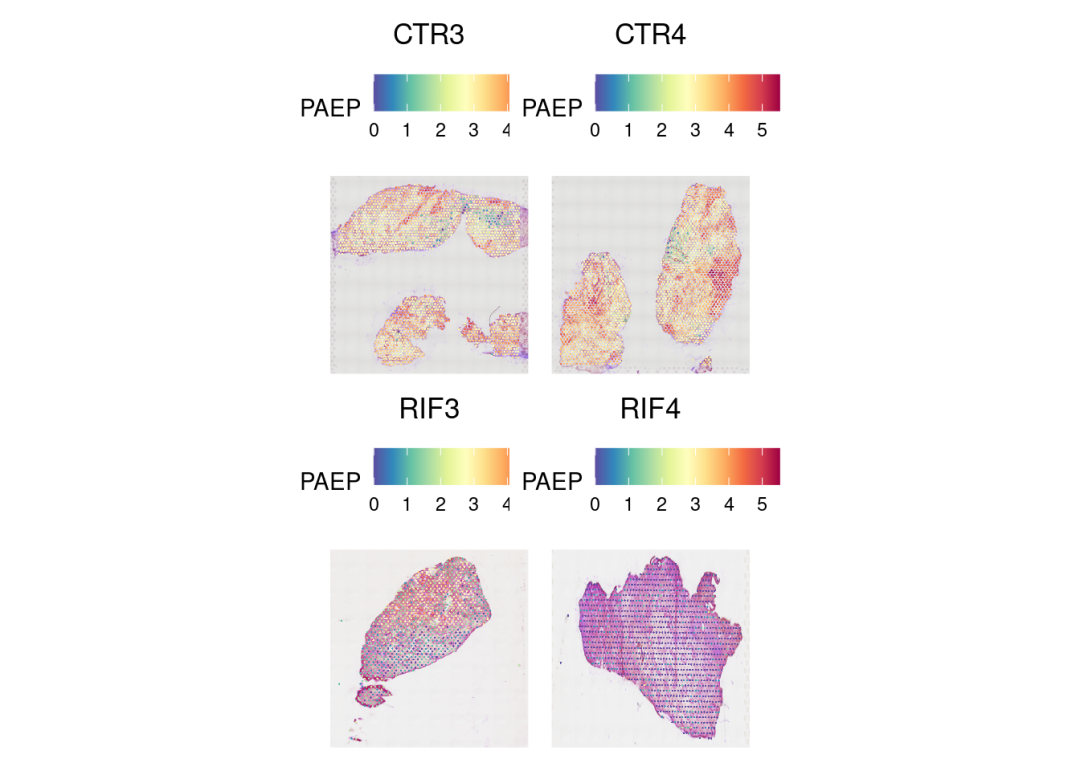
SpatialFeaturePlot(merged, 'SPP1', ncol = 2, min.cutoff = 0, stroke=0.1,pt.size.factor = 2)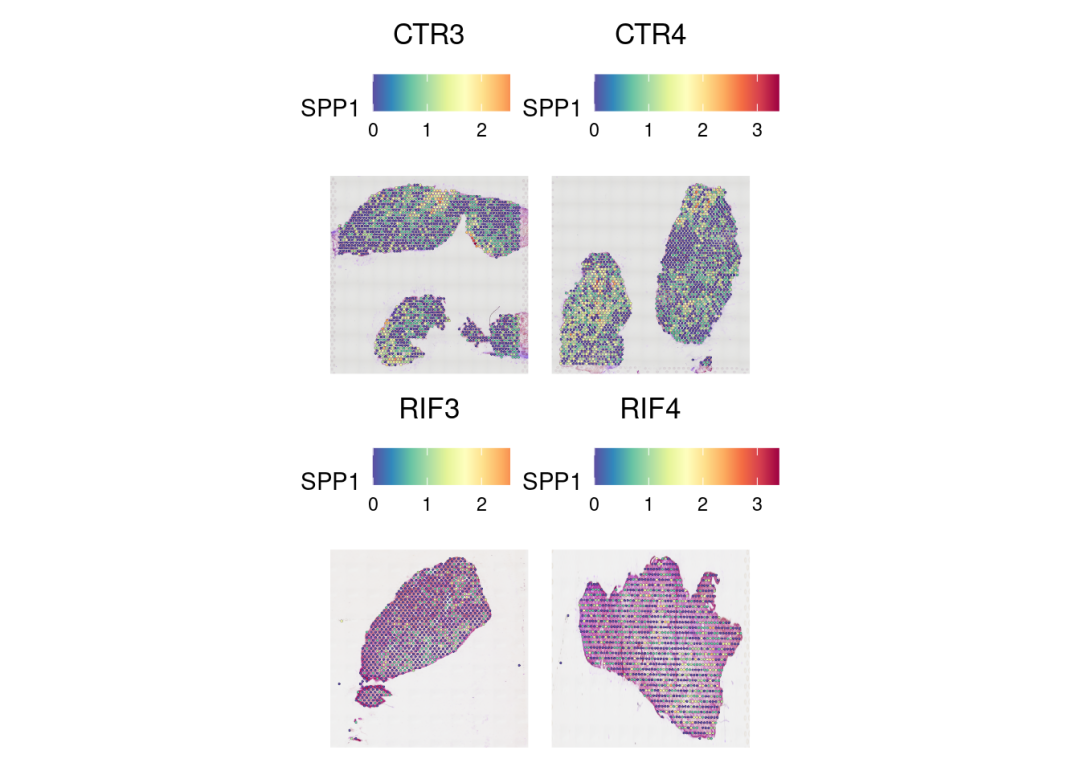
觉得分享有用的点个赞再走呗!下节继续反卷积的分析流程!
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-12-02,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

