听说infercnv不更新了?好在还能用!
听说infercnv不更新了?好在还能用!

KS科研分享与服务-TS的美梦
发布于 2026-01-22 13:49:28
发布于 2026-01-22 13:49:28
听说infercnv不更新了,所以上一期介绍了一款新的CNV分析工具(听名字就很快的单细胞CNV分析之fastCNV(谢绝运行时间焦虑))。infercnv是非常重要的分析工具,虽然不更新了,但是分析功能还正常!
Github:https://github.com/broadinstitute/infercnv
1、加载数据
library(infercnv)
library(Seurat)
library(SeuratObject)uterus <- readRDS("~/data_analysis/CNVs_analysis/uterus.rds")
uterus## An object of class Seurat
## 29001 features across 27914 samples within 2 assays
## Active assay: RNA (27001 features, 0 variable features)
## 2 layers present: counts, data
## 1 other assay present: integrated
## 2 dimensional reductions calculated: pca, tsne#我只选择上皮细胞-正常组和癌症EEC组进行分析
sce_CNV <- uterus[, Idents(uterus) %in% c("Unciliated epithelial cells","Ciliated epithelial cells")]
sce_CNV <- sce_CNV[,sce_CNV@meta.data$orig.ident %in% c("HC","EEC")]
sce_CNV## An object of class Seurat
## 29001 features across 2696 samples within 2 assays
## Active assay: RNA (27001 features, 0 variable features)
## 2 layers present: counts, data
## 1 other assay present: integrated
## 2 dimensional reductions calculated: pca, tsne2、准备分析文件
count Matrix:行为genes,列为cells的counts矩阵。
matrix_counts <- GetAssayData(sce_CNV,layer = 'counts',assay = 'RNA')
dim(matrix_counts)## [1] 27001 2696annotations_file:保存cell注释列为txt文件!一列是cells,一列是cellannotation。
#我们在metadata添加一列,将样本分组与celltype结合,区分正常细胞与肿瘤细胞
sce_CNV$newcelltype <- paste0(sce_CNV$orig.ident,"_",sce_CNV$celltype)
unique(sce_CNV$newcelltype)write.table(sce_CNV$newcelltype, "celltype.label.txt", sep = "\t", quote = F, col.names = F)annotations_file <- read.table('./celltype.label.txt', sep = "\t")
head(annotations_file)V1<chr> | V2<chr> | |
|---|---|---|
1 | AAACCCAAGTACGTCT-1_1 | HC_Unciliated epithelial cells |
2 | AAACCCAGTACCTATG-1_1 | HC_Ciliated epithelial cells |
3 | AAACCCATCCAAACCA-1_1 | HC_Unciliated epithelial cells |
4 | AAACCCATCTCGCAGG-1_1 | HC_Ciliated epithelial cells |
5 | AAACGAAAGACAGCTG-1_1 | HC_Ciliated epithelial cells |
6 | AAACGAAGTGGCTGCT-1_1 | HC_Unciliated epithelial cells |
gene_order_file:https://data.broadinstitute.org/Trinity/CTAT/cnv/.
hg38 <- read.table('./hg38_gencode_v27.txt',header = T)
head(hg38)DDX11L1<chr> | chr1<chr> | X11869<int> | X14409<int> | |
|---|---|---|---|---|
1 | WASH7P | chr1 | 14404 | 29570 |
2 | MIR6859-1 | chr1 | 17369 | 17436 |
3 | MIR1302-2HG | chr1 | 29554 | 31109 |
4 | MIR1302-2 | chr1 | 30366 | 30503 |
5 | FAM138A | chr1 | 34554 | 36081 |
6 | AL627309.6 | chr1 | 52473 | 53312 |
3、run infercnv
infercnv_obj = CreateInfercnvObject(raw_counts_matrix = matrix_counts,#count矩阵
annotations_file="./celltype.label.txt",#celltype信息
delim="\t",
gene_order_file="./hg38_gencode_v27.txt",
ref_group_names=c("HC_Unciliated epithelial cells",
"HC_Ciliated epithelial cells"),#HC组正常的细胞作为reference
chr_exclude=c("chrY","chrM"))#选择不需要的染色体,查看帮助函数去除## INFO [2026-01-08 22:54:51] Parsing gene order file: ./hg38_gencode_v27.txt
## INFO [2026-01-08 22:54:51] Parsing cell annotations file: ./celltype.label.txt
## INFO [2026-01-08 22:54:51] ::order_reduce:Start.
## INFO [2026-01-08 22:54:53] .order_reduce(): expr and order match.
## INFO [2026-01-08 22:54:55] ::process_data:order_reduce:Reduction from positional data, new dimensions (r,c) = 27001,2696 Total=31245803 Min=0 Max=2135.
## INFO [2026-01-08 22:54:55] num genes removed taking into account provided gene ordering list: 2159 = 7.99600014814266% removed.
## INFO [2026-01-08 22:54:55] -filtering out cells < 100 or > Inf, removing 0 % of cells
## WARN [2026-01-08 22:54:55] Please use "options(scipen = 100)" before running infercnv if you are using the analysis_mode="subclusters" option or you may encounter an error while the hclust is being generated.
## INFO [2026-01-08 22:54:56] validating infercnv_objinfercnv_obj = infercnv::run(infercnv_obj,
cutoff=0.1, # cutoff=1 works well for Smart-seq2, and cutoff=0.1 works well for 10x Genomics
out_dir="EEC", #分析结果out文件夹名
no_prelim_plot = T,
cluster_by_groups=T,
denoise=TRUE,
HMM=F,
min_cells_per_gene = 10,
num_threads=5,#线程数
write_expr_matrix = T#这里要选择T,分析后将结果exp矩阵导出,出现infercnv.observations.txt结果文件
)## INFO [2026-01-08 22:54:56] ::process_data:Start
## INFO [2026-01-08 22:54:56] Checking for saved results.
## INFO [2026-01-08 22:54:56] Trying to reload from step 22
## INFO [2026-01-08 22:54:59] Using backup from step 22
## INFO [2026-01-08 22:54:59] Trying to reload from step 15
## INFO [2026-01-08 22:55:08]
##
## STEP 1: incoming data
##
## INFO [2026-01-08 22:55:08]
##
## STEP 02: Removing lowly expressed genes
##
## INFO [2026-01-08 22:55:08]
##
## STEP 03: normalization by sequencing depth
##
## INFO [2026-01-08 22:55:08]
##
## STEP 04: log transformation of data
##
## INFO [2026-01-08 22:55:08]
##
## STEP 08: removing average of reference data (before smoothing)
##
## INFO [2026-01-08 22:55:08]
##
## STEP 09: apply max centered expression threshold: 3
##
## INFO [2026-01-08 22:55:08]
##
## STEP 10: Smoothing data per cell by chromosome
##
## INFO [2026-01-08 22:55:08]
##
## STEP 11: re-centering data across chromosome after smoothing
##
## INFO [2026-01-08 22:55:08]
##
## STEP 12: removing average of reference data (after smoothing)
##
## INFO [2026-01-08 22:55:08]
##
## STEP 14: invert log2(FC) to FC
##
## INFO [2026-01-08 22:55:08]
##
## STEP 15: computing tumor subclusters via leiden
##
## INFO [2026-01-08 22:55:08]
##
## STEP 22: Denoising
##
## INFO [2026-01-08 22:55:20]
##
## ## Making the final infercnv heatmap ##
## INFO [2026-01-08 22:55:21] ::plot_cnv:Start
## INFO [2026-01-08 22:55:21] ::plot_cnv:Current data dimensions (r,c)=11113,2696 Total=30092807.2516359 Min=0.736331334009685 Max=1.79721424399281.
## INFO [2026-01-08 22:55:22] ::plot_cnv:Depending on the size of the matrix this may take a moment.
## INFO [2026-01-08 22:56:07] plot_cnv(): auto thresholding at: (0.832440 , 1.167560)
## INFO [2026-01-08 22:56:08] plot_cnv_observation:Start
## INFO [2026-01-08 22:56:08] Observation data size: Cells= 1036 Genes= 11113
## INFO [2026-01-08 22:56:09] plot_cnv_observation:Writing observation groupings/color.
## INFO [2026-01-08 22:56:09] plot_cnv_observation:Done writing observation groupings/color.
## INFO [2026-01-08 22:56:09] plot_cnv_observation:Writing observation heatmap thresholds.
## INFO [2026-01-08 22:56:09] plot_cnv_observation:Done writing observation heatmap thresholds.## INFO [2026-01-08 22:56:15] Colors for breaks: #00008B,#24249B,#4848AB,#6D6DBC,#9191CC,#B6B6DD,#DADAEE,#FFFFFF,#EEDADA,#DDB6B6,#CC9191,#BC6D6D,#AB4848,#9B2424,#8B0000
## INFO [2026-01-08 22:56:15] Quantiles of plotted data range: 0.832440420996389,1.00134184739381,1.00134184739381,1.00134184739381,1.16755957900361## INFO [2026-01-08 22:56:18] plot_cnv_observations:Writing observation data to EEC/infercnv.observations.txt
## INFO [2026-01-08 22:56:35] plot_cnv_references:Start
## INFO [2026-01-08 22:56:35] Reference data size: Cells= 1660 Genes= 11113
## INFO [2026-01-08 22:57:30] plot_cnv_references:Number reference groups= 2
## INFO [2026-01-08 22:57:30] plot_cnv_references:Plotting heatmap.## INFO [2026-01-08 22:57:38] Colors for breaks: #00008B,#24249B,#4848AB,#6D6DBC,#9191CC,#B6B6DD,#DADAEE,#FFFFFF,#EEDADA,#DDB6B6,#CC9191,#BC6D6D,#AB4848,#9B2424,#8B0000
## INFO [2026-01-08 22:57:38] Quantiles of plotted data range: 0.832440420996389,1.00134184739381,1.00134184739381,1.00134184739381,1.16755957900361## INFO [2026-01-08 22:57:40] plot_cnv_references:Writing reference data to EEC/infercnv.references.txtlibrary(RColorBrewer)
infercnv::plot_cnv(infercnv_obj,
output_filename = "inferCNV_heatmap",
output_format = "pdf",
custom_color_pal = color.palette(c("#2067AE","white","#B11F2B"))) #改颜色## INFO [2026-01-08 22:58:07] ::plot_cnv:Start
## INFO [2026-01-08 22:58:07] ::plot_cnv:Current data dimensions (r,c)=11113,2696 Total=30092807.2516359 Min=0.736331334009685 Max=1.79721424399281.
## INFO [2026-01-08 22:58:07] ::plot_cnv:Depending on the size of the matrix this may take a moment.
## INFO [2026-01-08 22:58:09] plot_cnv(): auto thresholding at: (0.841263 , 1.167560)
## INFO [2026-01-08 22:58:11] plot_cnv_observation:Start
## INFO [2026-01-08 22:58:11] Observation data size: Cells= 1036 Genes= 11113
## INFO [2026-01-08 22:58:11] plot_cnv_observation:Writing observation groupings/color.
## INFO [2026-01-08 22:58:11] plot_cnv_observation:Done writing observation groupings/color.
## INFO [2026-01-08 22:58:11] plot_cnv_observation:Writing observation heatmap thresholds.
## INFO [2026-01-08 22:58:11] plot_cnv_observation:Done writing observation heatmap thresholds.## INFO [2026-01-08 22:58:16] Colors for breaks: #2067AE,#3F7CB9,#5F92C5,#7FA8D0,#9FBDDC,#BFD3E7,#DFE9F3,#FFFFFF,#F3DFE0,#E8BFC2,#DD9FA4,#D27F85,#C75F67,#BC3F49,#B11F2B
## INFO [2026-01-08 22:58:16] Quantiles of plotted data range: 0.841262610131915,1.00134184739381,1.00134184739381,1.00134184739381,1.16755957900361## INFO [2026-01-08 22:58:18] plot_cnv_references:Start
## INFO [2026-01-08 22:58:18] Reference data size: Cells= 1660 Genes= 11113
## INFO [2026-01-08 22:59:12] plot_cnv_references:Number reference groups= 2
## INFO [2026-01-08 22:59:13] plot_cnv_references:Plotting heatmap.## INFO [2026-01-08 22:59:20] Colors for breaks: #2067AE,#3F7CB9,#5F92C5,#7FA8D0,#9FBDDC,#BFD3E7,#DFE9F3,#FFFFFF,#F3DFE0,#E8BFC2,#DD9FA4,#D27F85,#C75F67,#BC3F49,#B11F2B
## INFO [2026-01-08 22:59:20] Quantiles of plotted data range: 0.841262610131915,1.00134184739381,1.00134184739381,1.00134184739381,1.16755957900361## $cluster_by_groups
## [1] TRUE
##
## $k_obs_groups
## [1] 1
##
## $contig_cex
## [1] 1
##
## $x.center
## [1] 1.004411
##
## $x.range
## [1] 0.8412626 1.1675596
##
## $hclust_method
## [1] "ward.D"
##
## $color_safe_pal
## [1] FALSE
##
## $output_format
## [1] "pdf"
##
## $png_res
## [1] 300
##
## $dynamic_resize
## [1] 0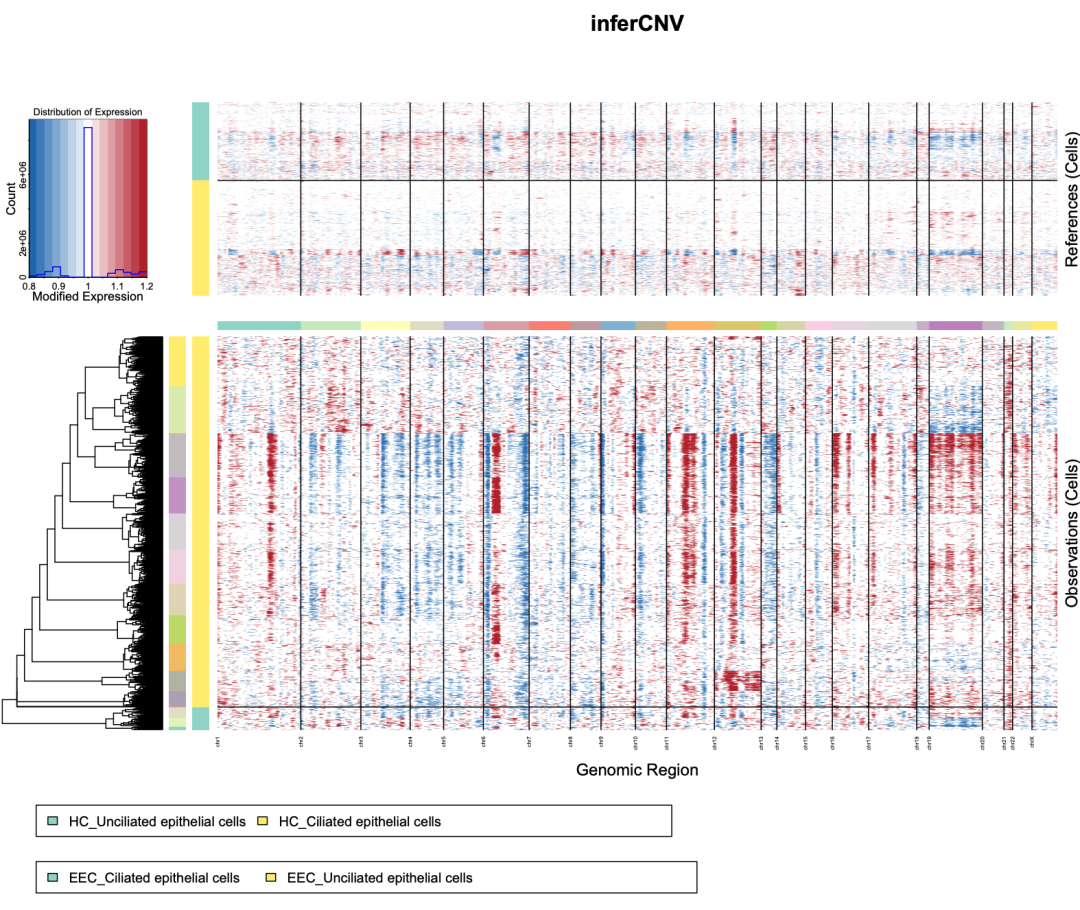
觉得分享有用的点个赞再走呗,祝您心想事成,文章顺利,毕业顺利!
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2026-01-22,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

