分子对接PyRasetta--Protein Geometry
原创分子对接PyRasetta--Protein Geometry
原创
追风少年i
发布于 2025-09-22 10:46:37
发布于 2025-09-22 10:46:37
代码可运行
运行总次数:0
代码可运行
作者,Evil Genius
今天我们继续分子对接,有几个问题大家自己要学习一下,首先要认识Cα原子。
简单补充一下chi1,phi, psi。
chi1(chi-one)、phi 和 psi 是用于描述蛋白质分子中氨基酸残基构象的二面角。
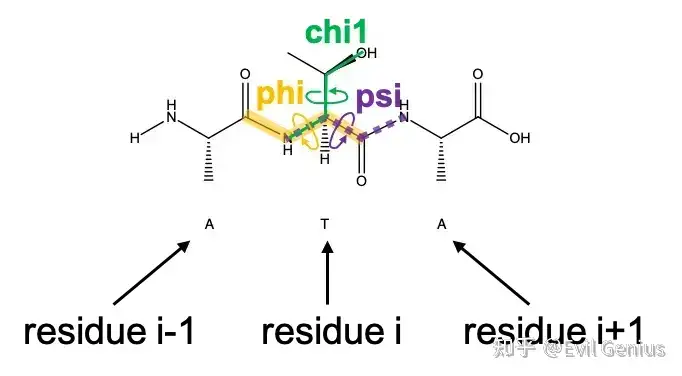
主链二面角:phi 和 psi
phi 和 psi 专门用来描述蛋白质主链(或骨架) 的构象。蛋白质的主链是由重复的 -N-Cα-C- 单元组成的。
phi角 (φ):
定义:围绕 N-Cα 键 的旋转角。
涉及的四个原子: C'(i-1) - N(i) - Cα(i) - C'(i)
(i) 代表第 i 个氨基酸残基。
作用: 主要决定了主链在氨基酸氨基(N-H)一侧的走向。
psi角 (ψ):
定义:围绕 Cα-C 键 的旋转角。
涉及的四个原子: N(i) - Cα(i) - C'(i) - N(i+1)
作用: 主要决定了主链在氨基酸羧基(C=O)一侧的走向。
为什么 phi 和 psi 如此重要?
这两个角度的组合直接决定了蛋白质主链的二级结构,例如:
α-螺旋: 有一套特征性的、重复的 phi 和 psi 值(大约为 -60°, -45°)。
β-折叠: 有另一套特征性的 phi 和 psi 值(大约为 -120°, 120°)。
通过绘制所有氨基酸残基的 phi 和 psi 值,可以生成著名的 拉氏图,用于评估蛋白质模型的构象是否合理。
侧链二面角:chi1
chi1 是用于描述氨基酸侧链构象的二面角。蛋白质的20种常见氨基酸有不同数量的侧链二面角,用 chi1, chi2, chi3... 表示。chi1 是第一个,也是最关键的一个。
定义:围绕 N-Cα 键 的旋转角。
涉及的四个原子:对于大多数氨基酸(除甘氨酸、丙氨酸等侧链简单的以外),chi1 定义为:
N(i) - Cα(i) - Cβ(i) - X(i)
X 是侧链上的下一个原子。对于缬氨酸、半胱氨酸等是 Cγ,对于丝氨酸是 Oγ,等等。
作用: chi1 决定了侧链从主链上“伸出去”的方向,是朝向分子内部还是朝向溶剂。它对于侧链的堆积、形成疏水核心以及与其他分子相互作用至关重要。
然后我们继续,蛋白几何结构
Keywords: pose_from_sequence(), bond_angle(), set_phi(), set_psi(), xyz()
!pip install pyrosettacolabsetup
import pyrosettacolabsetup; pyrosettacolabsetup.install_pyrosetta()
import pyrosetta; pyrosetta.init()
from pyrosetta import *
from pyrosetta.teaching import *
init()
pose = pose_from_pdb("inputs/5tj3.pdb")
resid = pose.pdb_info().pdb2pose('A', 28)
res_28 = pose.residue(resid)
N28 = AtomID(res_28.atom_index("N"), resid)
CA28 = AtomID(res_28.atom_index("CA"), resid)
C28 = AtomID(res_28.atom_index("C"), resid)查看一下Rosetta为该氨基酸键长设定的理想值,并比较这些数值。
from IPython.display import Image
Image('./Media/init-path.png',width='700')通过 ICOOR_INTERNAL 数据行可以获取该残基中相邻原子间的二面角、键角以及键长信息。由此应能推断出 Rosetta 中设定的 N-Cα 和 Cα-C 键理想键长值。
one_res_seq = "V"
pose_one_res = pose_from_sequence(one_res_seq)
print(pose_one_res.sequence())
N_xyz = pose_one_res.residue(1).xyz("N")
CA_xyz = pose_one_res.residue(1).xyz("CA")
C_xyz = pose_one_res.residue(1).xyz("C")
print((CA_xyz - N_xyz).norm())
print((CA_xyz - C_xyz).norm())
1.458004
1.52326如何获取蛋白质中的键角信息。如果Conformation类中包含我们所需的键角数据,可以使用之前已经创建的AtomID对象来实现:
angle = pose.conformation().bond_angle(N28, CA28, C28)
print(angle)
1.913188766577386
import math
angle*180/math.pi
109.61764173672383该数值与基于Cα碳原子四面体几何结构所预期角度的对比结果。
Exercise 5: Calculating psi angle(我们做一下这个练习,大家自己也动手做一下)。
Try to calculate this angle using the xyz atom positions for N, CA, and C of residue A:28 in the protein. You can use the Vector function v3 = v1.dot(v2) along with v1.norm(). The vector angle between two vectors BA and BC is cos−1(BA⋅BC|BA||BC|)
# 导入PyRosetta
import pyrosetta
from pyrosetta import rosetta
import math
# 确保已经初始化
pyrosetta.init()
# 假设pose已经包含你的蛋白质结构
# pose = pyrosetta.pose_from_pdb("your_protein.pdb")
# 定义要计算psi角的残基序号
resid = 28 # 残基A:28
# 获取指定残基的N, CA, C原子的xyz坐标
# 注意:Rosetta中原子索引是固定的,N=1, CA=2, C=3
N = pose.residue(resid).xyz("N")
CA = pose.residue(resid).xyz("CA")
C = pose.residue(resid).xyz("C")
# 获取下一个残基的N原子坐标(对于psi角计算需要)
N_next = pose.residue(resid + 1).xyz("N")
# 计算三个关键向量
# 向量1: CA -> N
vector_CA_to_N = N - CA
# 向量2: CA -> C
vector_CA_to_C = C - CA
# 向量3: C -> N_next (下一个残基的N)
vector_C_to_N_next = N_next - C
# 计算二面角psi所需的四个平面
# psi角定义为: N(i)-CA(i)-C(i)-N(i+1) 的二面角
# 计算两个法向量
# 平面1的法向量 (N-CA-C)
vector1 = vector_CA_to_N
vector2 = vector_CA_to_C
normal1 = vector1.cross(vector2)
# 平面2的法向量 (CA-C-N_next)
vector3 = -vector_CA_to_C # C->CA 的反向向量
vector4 = vector_C_to_N_next
normal2 = vector3.cross(vector4)
# 计算psi角
# 使用点积公式计算二面角
cos_psi = normal1.dot(normal2) / (normal1.norm() * normal2.norm())
psi_rad = math.acos(cos_psi)
# 确定二面角符号(使用右手法则)
if normal1.dot(vector4) < 0:
psi_rad = -psi_rad
# 转换为角度
psi_deg = math.degrees(psi_rad)
print(f"Residue A:{resid} 的psi角: {psi_deg:.2f}°")
# 更简单的方法:使用PyRosetta内置函数验证
# Rosetta可以直接计算任何二面角
calculated_psi = pose.psi(resid)
print(f"使用PyRosetta内置函数验证: {calculated_psi:.2f}°")操纵蛋白质几何结构
同样可以改变蛋白质的几何构型,特别关注于操纵蛋白质主链和χ二面角。
更改phi/psi角
# three alanines
tripeptide = pose_from_sequence("AAA")
orig_phi = tripeptide.phi(2)
orig_psi = tripeptide.psi(2)
print("original phi:", orig_phi)
print("original psi:", orig_psi)
# print the xyz coordinates of the CB atom of residue 2 here BEFORE setting
### BEGIN SOLUTION
print("xyz coordinates:", tripeptide.residue(2).xyz("CB"))
### END SOLUTION
original phi: 180.0
original psi: 180.0
xyz coordinates: 3.535270304899897 3.659035776744378 1.199094204197625
# set the phi and psi here
### BEGIN SOLUTION
tripeptide.set_phi(2, -60)
tripeptide.set_psi(2, -43)
print("new phi:", tripeptide.phi(2))
print("new psi:", tripeptide.psi(2))
### END SOLUTION
# print the xyz coordinates of the CB atom of residue 2 here AFTER setting
### BEGIN SOLUTION
print("xyz coordinates:", tripeptide.residue(2).xyz("CB"))
### END SOLUTION
# did changing the phi and psi angle change the xyz coordinates of the CB atom of alanine 2?
xyz coordinates: 5.498535447297188 2.671616235800968 0.06968570437453583FOLD_TREE(折叠树)控制着残基几何结构的改变如何沿着蛋白质传递(沿FoldTree链从左向右传播)。
print(pose)
PDB file name: 5tj3.pdb
Total residues:524
Sequence: NAVPRPKLVVGLVVDQMRWDYLYRYYSKYGEGGFKRMLNTGYSLNNVHIDYVPTVTAIGHTSIFTGSVPSIHGIAGNDWYDKELGKSVYCTSDETVQPVGTTSNSVGQHSPRNLWSTTVTDQLGLATNFTSKVVGVSLKDRASILPAGHNPTGAFWFDDTTGKFITSTYYTKELPKWVNDFNNKNVPAQLVANGWNTLLPINQYTESSEDNVEWEGLLGSKKTPTFPYTDLAKDYEAKKGLIRTTPFGNTLTLQMADAAIDGNQMGVDDITDFLTVNLASTDYVGHNFGPNSIEVEDTYLRLDRDLADFFNNLDKKVGKGNYLVFLSADHGAAHSVGFMQAHKMPTGFFDMKKEMNAKLKQKFGADNIIAAAMNYQVYFDRKVLADSKLELDDVRDYVMTELKKEPSVLYVLSTDEIWESSIPEPIKSRVINGYNWKRSGDIQIISKDGYLSAYSKKGTTHSVWNSYDSHIPLLFMGWGIKQGESNQPYHMTDIAPTVSSLLKIQFPSGAVGKPITEVIGZZZZ
Fold tree:
FOLD_TREE EDGE 1 520 -1 EDGE 1 521 1 EDGE 1 522 2 EDGE 1 523 3 EDGE 1 524 4今天我们就到这里,生活很好,有你更好
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录

