ggcoverage优雅的绘制各种组学注释图
欢迎关注R语言数据分析指南
❝本节来介绍如何使用「ggcoverage」来对各种测序数据进行分析,包括WGS、RNA-seq、ChIP-seq、ATAC-seq等,下面小编来简单介绍下,更多详细的案例内容请参考作者官方文档。 ❞
官方文档
❝https://showteeth.github.io/ggcoverage/ ❞加载R包
BiocManager::install("areyesq89/GenomeMatrix")
remotes::install_github("showteeth/ggcoverage")
library("rtracklayer")
library("ggcoverage")
library("ggpattern")
导入内部数据
meta.file <- system.file("extdata", "RNA-seq", "meta_info.csv", package = "ggcoverage")
sample.meta = read.csv(meta.file)
track.folder = system.file("extdata", "RNA-seq", package = "ggcoverage")
track.df = LoadTrackFile(track.folder = track.folder, format = "bw",
region = "chr14:21,677,306-21,737,601", extend = 2000,
meta.info = sample.meta)
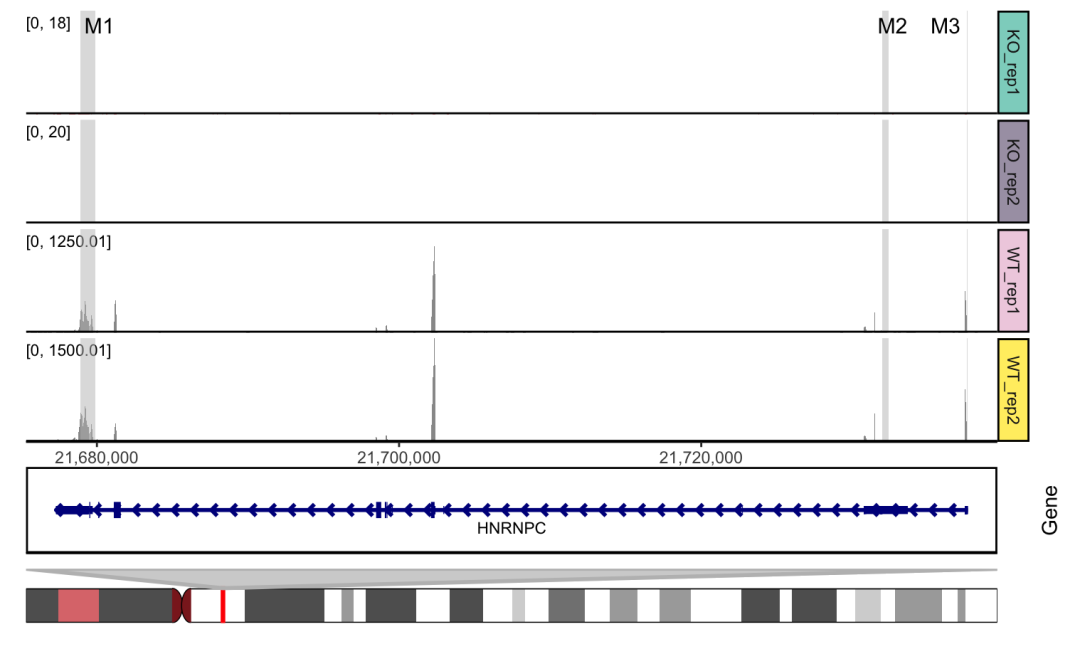
准备标记区域
mark.region=data.frame(start=c(21678900,21732001,21737590),
end=c(21679900,21732400,21737650),
label=c("M1", "M2", "M3"))
加载gff文件
gtf.file = system.file("extdata", "used_hg19.gtf", package = "ggcoverage")
gtf.gr = rtracklayer::import.gff(con = gtf.file, format = 'gtf')
添加基因注释
basic.coverage = ggcoverage(data = track.df, color = "auto", plot.type = "facet",
mark.region = mark.region, range.position = "in",
facet.y.scale = "fixed")
basic.coverage + geom_gene(gtf.gr=gtf.gr)

basic.coverage +
geom_gene(gtf.gr=gtf.gr) +
geom_ideogram(genome = "hg19",plot.space = 0)
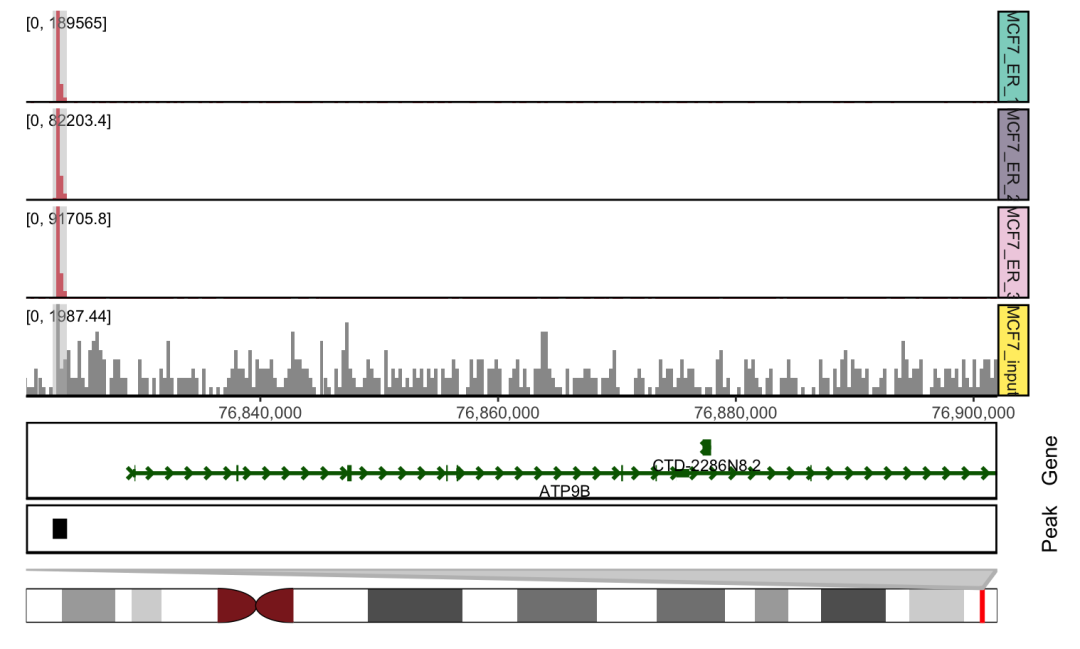
ChIP-seq案例展示
sample.meta = data.frame(SampleName=c('Chr18_MCF7_ER_1','Chr18_MCF7_ER_2','Chr18_MCF7_ER_3','Chr18_MCF7_input'),
Type = c("MCF7_ER_1","MCF7_ER_2","MCF7_ER_3","MCF7_input"),
Group = c("IP", "IP", "IP", "Input"))
track.folder = system.file("extdata", "ChIP-seq", package = "ggcoverage")
track.df = LoadTrackFile(track.folder = track.folder, format = "bw", region = "chr18:76822285-76900000",
meta.info = sample.meta)
mark.region=data.frame(start=c(76822533),end=c(76823743),label=c("Promoter"))
basic.coverage = ggcoverage(data = track.df, color = "auto",
mark.region=mark.region, show.mark.label = FALSE)
peak.file = system.file("extdata", "ChIP-seq", "consensus.peak", package = "ggcoverage")
basic.coverage +
geom_gene(gtf.gr=gtf.gr) +
geom_peak(bed.file = peak.file) +
geom_ideogram(genome = "hg19",plot.space = 0)
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2023-08-12,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录
