多组差异表达分析的可视化
多组差异表达分析的可视化

DoubleHelix
发布于 2023-09-06 10:04:52
发布于 2023-09-06 10:04:52
代码可运行
运行总次数:0
代码可运行
degr = dir("output/016_hot_cold_tumor/DEG",
"DESeq2-filtered.csv$",full.names = T,recursive = T)
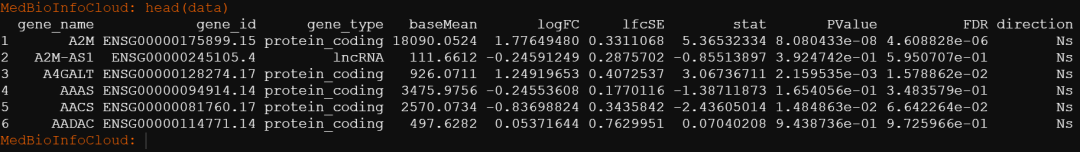
degr[1:4]MedBioInfoCloud: degr[1:4]
[1] "output/016_hot_cold_tumor/DEG/TCGA-ACC/DESeq2-filtered.csv"
[2] "output/016_hot_cold_tumor/DEG/TCGA-BLCA/DESeq2-filtered.csv"
[3] "output/016_hot_cold_tumor/DEG/TCGA-BRCA/DESeq2-filtered.csv"
[4] "output/016_hot_cold_tumor/DEG/TCGA-CESC/DESeq2-filtered.csv"读入其中一个:
data = read.csv(degr[1],header = T,
check.names = F,row.names = 1)查看一下数据:
我这里的差异分析结果文件很多,我选择4个文件读入并合并。
subdeg = degr[1:4]
alldeg = do.call(rbind,lapply(subdeg, function(x){
data = read.csv(x,header = T,
check.names = F,row.names = 1)
data = data[data$gene_type == "protein_coding",]
data = data[!duplicated(data[,"gene_name"]),]
data$cancer = unlist(strsplit(x,"/"))[4]
data$cancer = unlist(strsplit(data$cancer,"-"))[2]
return(data)
}))合并后添加了1列cancer。
处理一下数据,并增加一列cluster
head(alldeg)
alldeg2 = alldeg[alldeg$direction != "Ns",]
alldeg2 = arrange(alldeg2,cancer)
alldeg2$cancer = factor(alldeg2$cancer)
alldeg2$cluster = as.numeric(alldeg2$cancer) - 1MedBioInfoCloud: head(alldeg2)[,(ncol(alldeg2)-3):ncol(alldeg2)]
FDR direction cancer cluster
27 1.164291e-06 Up ACC 0
33 1.452798e-09 Down ACC 0
182 9.039400e-04 Up ACC 0
430 4.099443e-03 Down ACC 0
447 4.636853e-11 Down ACC 0
469 8.875275e-05 Up ACC 0计算每组差异分析中logFC的最大值和最小值
minlogfc = alldeg2 %>% group_by(cancer) %>% dplyr::slice(which.min(logFC))
maxlogfc = alldeg2 %>% group_by(cancer) %>% dplyr::slice(which.max(logFC))
根据分组个数,定义用来绘图的数据。
dfbar0 <- data.frame(x=c(0:3),
y= maxlogfc$logFC )
dfbar1<- data.frame(x=c(0:3),
y=minlogfc$logFC)
dfcol<- data.frame(x=c(0:3),
y=0,
label= unique(alldeg2$cancer))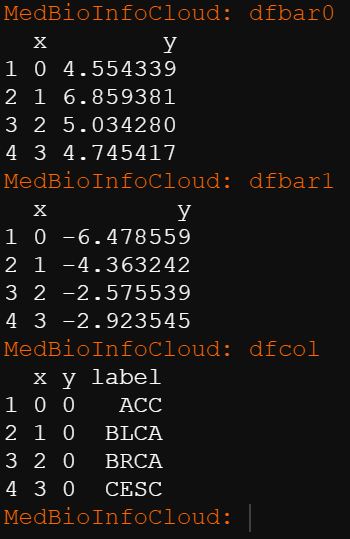
绘制背景图:
p <- ggplot()+
geom_col(data = dfbar0,
mapping = aes(x = x,y = y),
fill = "#dcdcdc",alpha = 0.6)+
geom_col(data = dfbar1,
mapping = aes(x = x,y = y),
fill = "#dcdcdc",alpha = 0.6) 
添加散点图:
p1 = p + geom_jitter(data = alldeg2,
aes(x = cluster, y = logFC, color = direction),
size = 0.85,
width =0.4)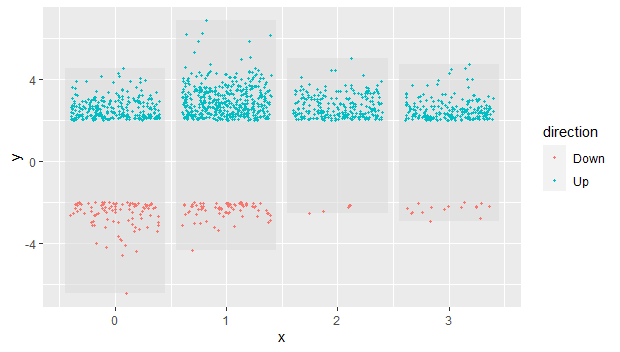
修改点的颜色:
p2 = p1+ scale_color_manual(name=NULL,
values = c("blue","red"))
添加注释框:
p3 = p2 + geom_tile(data = dfcol,
aes(x=x,y=y),
height=2,
color = "black",
fill = mycol,
alpha = 0.6,
show.legend = F)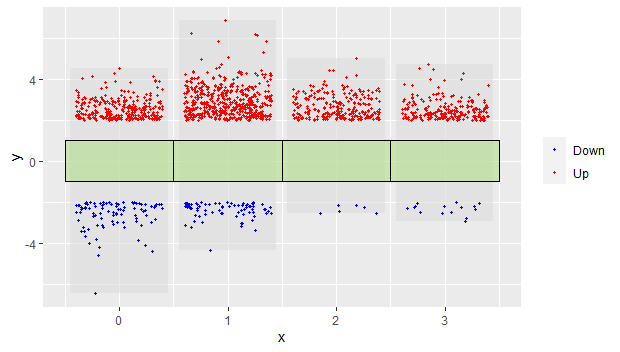
添加文本和坐标标题:
p4 = p3 +
labs(x="Cancer",y="log2FC") + #添加坐标标题
##给注释框添加文本
geom_text(data=dfcol,
aes(x=x,y=y,label=label),
size =6,
color ="black")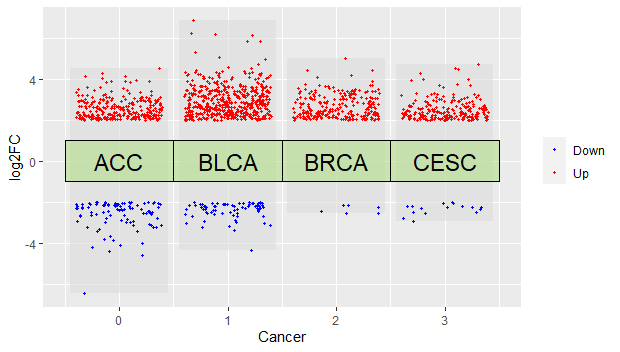
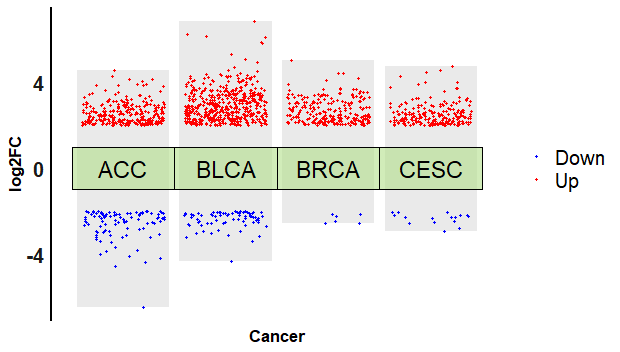
修改主题,需要把横坐标的数值去掉,因为它没有任何意义。
p4 + theme_minimal()+
theme(
axis.title = element_text(size = 13,
color = "black",
face = "bold"),
axis.line.y = element_line(color = "black",
size = 1),
axis.line.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 15,face = "bold", colour = "#1A1A1A"),
panel.grid = element_blank(),
legend.direction = "vertical",
legend.text = element_text(size = 15)
)
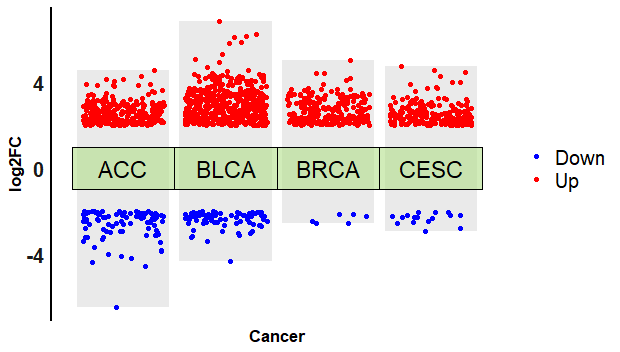
完整代码:
ggplot()+
geom_col(data = dfbar0,
mapping = aes(x = x,y = y),
fill = "#dcdcdc",alpha = 0.6)+
geom_col(data = dfbar1,
mapping = aes(x = x,y = y),
fill = "#dcdcdc",alpha = 0.6)+
geom_jitter(data = alldeg2,
aes(x = cluster, y = logFC, color = direction),
size = 1.5,
width =0.4)+
scale_color_manual(name=NULL,
values = c("blue","red"))+
geom_tile(data = dfcol,
aes(x=x,y=y),
height=2,
color = "black",
fill = mycol,
alpha = 0.6,
show.legend = F)+
labs(x="Cancer",y="log2FC") + #添加坐标标题
##给注释框添加文本
geom_text(data=dfcol,
aes(x=x,y=y,label=label),
size =6,
color ="black")+
theme_minimal()+
theme(
axis.title = element_text(size = 13,
color = "black",
face = "bold"),
axis.line.y = element_line(color = "black",
size = 1),
axis.line.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 15,face = "bold", colour = "#1A1A1A"),
panel.grid = element_blank(),
legend.direction = "vertical",
legend.text = element_text(size = 15)
)
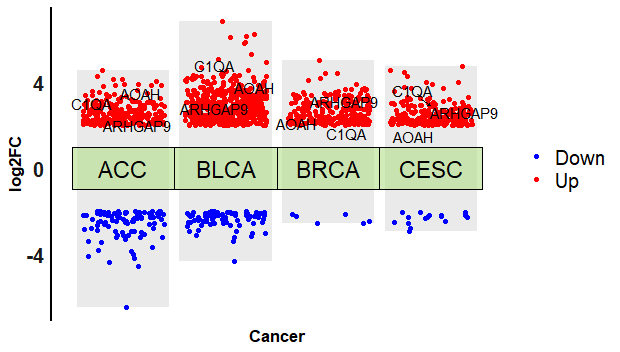
统计所有分组差异基因的频率,找出共同基因并标注出来。
gene_stat = as.data.frame(table(alldeg2$gene_name))
gene_stat = arrange(gene_stat,desc(Freq))
head(gene_stat)MedBioInfoCloud: head(gene_stat)
Var1 Freq
1 AOAH 4
2 ARHGAP9 4
3 C1QA 4
4 C1QB 4
5 C1QC 4总共4个分组的差异分析,频率为4的基因就是共同的差异表达基因。我们选择3个来显示:
gs = gene_stat$Var1[gene_stat$Freq ==4][1:3]
gstab = alldeg2[alldeg2$gene_name %in% gs,]
gstab = arrange(gstab,cancer)library(ggrepel)
fig +
geom_text_repel(
data=gstab,
aes(x=cluster,y=logFC,label=gene_name),
force = 1.2,
arrow = arrow(length = unit(0.008, "npc"),
type = "open", ends = "last")
)
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2023-06-23,如有侵权请联系 cloudcommunity@tencent.com 删除
本文分享自 MedBioInfoCloud 微信公众号,前往查看
如有侵权,请联系 cloudcommunity@tencent.com 删除。
本文参与 腾讯云自媒体同步曝光计划 ,欢迎热爱写作的你一起参与!
评论
登录后参与评论
推荐阅读

