ggpicrust2优雅的探索PICRUSt2分析数据
ggpicrust2优雅的探索PICRUSt2分析数据
R语言数据分析指南
发布于 2023-09-11 08:43:12
发布于 2023-09-11 08:43:12
代码可运行
运行总次数:0
代码可运行
欢迎关注R语言数据分析指南
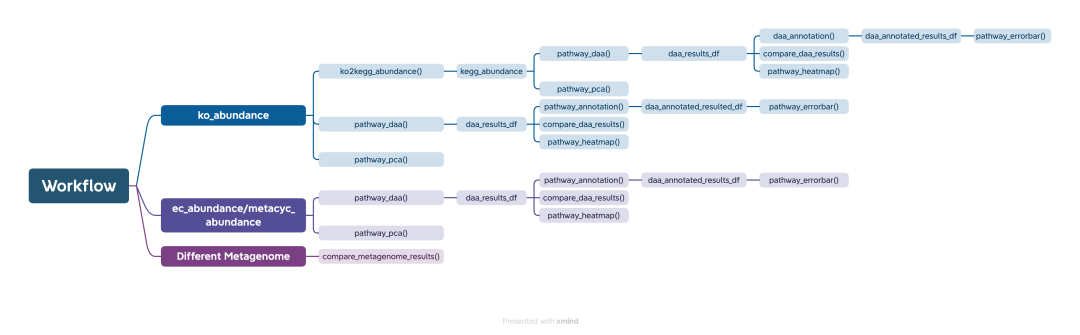
❝本节来介绍一款R包「ggpicrust2」,主要用于对「PICRUSt2」输出的结果进行深度操作,ggpicrust2集成了ko丰度到kegg通路丰度转换、通路注释、差异丰度(DA)分析及结果图的可视化等。下面小编来简单介绍下,更多详细的案例内容请参考作者官方文档。「此款R包安装依赖较多,请耐心安装」 ❞
官方文档
❝https://github.com/cafferychen777/ggpicrust2 ❞
安装R包
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
pkgs <- c("phyloseq", "ALDEx2", "SummarizedExperiment", "Biobase", "devtools",
"ComplexHeatmap", "BiocGenerics", "BiocManager", "metagenomeSeq",
"Maaslin2", "edgeR", "lefser", "limma", "KEGGREST", "DESeq2")
for (pkg in pkgs) {
if (!requireNamespace(pkg, quietly = TRUE))
BiocManager::install(pkg)
}
devtools::install_github("cafferychen777/ggpicrust2")
加载R包
library(ggpicrust2)
library(tidyverse)
library(GGally)
library(ggprism)
library(patchwork)
library(ggh4x)
差异丰度分析
data("metacyc_abundance")
data("metadata")
metacyc_daa_results_df <- pathway_daa(abundance = metacyc_abundance %>%
column_to_rownames("pathway"), metadata = metadata,
group = "Environment", daa_method = "LinDA",
select = NULL, p.adjust = "BH", reference = NULL)
多模型结果比较
methods <- c("ALDEx2", "DESeq2", "edgeR")
daa_results_list <- lapply(methods, function(method) {
pathway_daa(abundance = metacyc_abundance %>%
column_to_rownames("pathway"), metadata = metadata, group = "Environment", daa_method = method)
})
method_names <- c("ALDEx2_Welch's t test","ALDEx2_Wilcoxon rank test","DESeq2", "edgeR")
# Compare results across different methods
comparison_results <- compare_daa_results(daa_results_list = daa_results_list, method_names = method_names)
> comparison_results
method num_features num_common_features num_diff_features
1 ALDEx2_Welch's t test 124 10 124
2 ALDEx2_Wilcoxon rank test 41 10 41
3 DESeq2 41 10 41
kegg通路注释
data("kegg_abundance")
data("metadata")
daa_results_df <- pathway_daa(abundance = kegg_abundance,
metadata = metadata, group = "Environment", daa_method = "LinDA")
daa_annotated_results_df <- pathway_annotation(pathway = "KO",
daa_results_df = daa_results_df, ko_to_kegg = TRUE)
Ko号注释
data("ko_abundance")
data("metadata")
daa_results_df <- pathway_daa(abundance = ko_abundance %>%
column_to_rownames("#NAME"), metadata = metadata,
group = "Environment", daa_method = "LinDA")
daa_annotated_results_df <- pathway_annotation(pathway = "KO",
daa_results_df = daa_results_df, ko_to_kegg = FALSE)
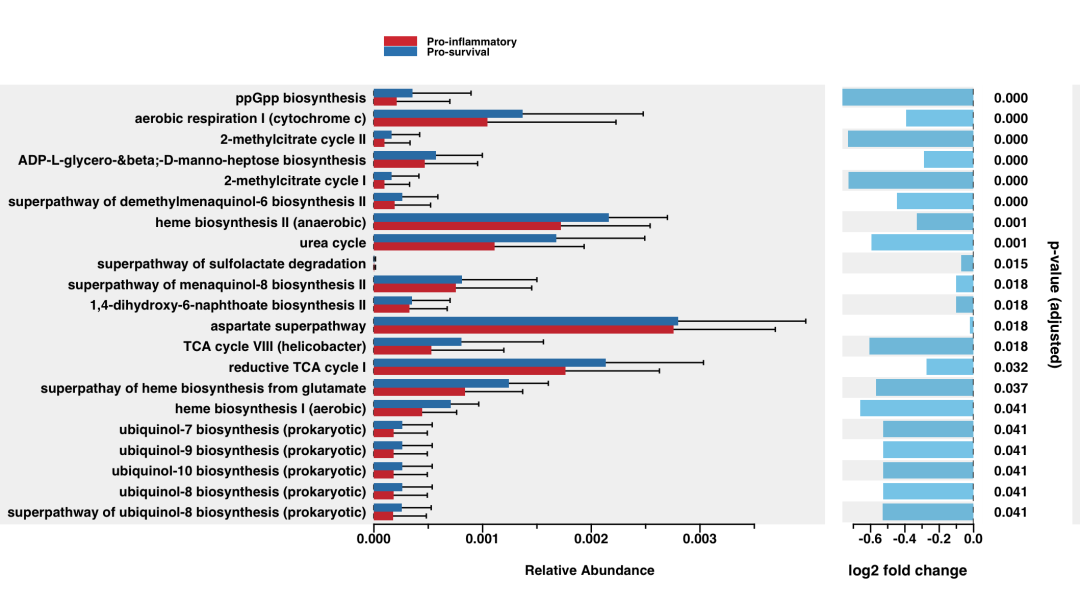
pathway_errorbar
data("ko_abundance")
data("metadata")
kegg_abundance <- ko2kegg_abundance(data = ko_abundance)
daa_results_df <- pathway_daa(kegg_abundance, metadata = metadata,
group = "Environment", daa_method = "LinDA")
daa_annotated_results_df <- pathway_annotation(pathway = "KO",
daa_results_df = daa_results_df, ko_to_kegg = TRUE)
p <- pathway_errorbar(abundance = kegg_abundance,
daa_results_df = daa_annotated_results_df,
Group = metadata$Environment,
ko_to_kegg = TRUE,
p_values_threshold = 0.05,
order = "pathway_class",
select = NULL,
p_value_bar = TRUE,
colors = NULL,
x_lab = "pathway_name")

data("metacyc_abundance")
data("metadata")
metacyc_daa_results_df <- pathway_daa(abundance = metacyc_abundance %>%
column_to_rownames("pathway"), metadata = metadata,
group = "Environment", daa_method = "LinDA")
metacyc_daa_annotated_results_df <- pathway_annotation(pathway = "MetaCyc",
daa_results_df = metacyc_daa_results_df, ko_to_kegg = FALSE)
p <- pathway_errorbar(abundance = metacyc_abundance %>%
column_to_rownames("pathway"),
daa_results_df = metacyc_daa_annotated_results_df,
Group = metadata$Environment,
ko_to_kegg = FALSE,
p_values_threshold = 0.05,
order = "group",
select = NULL,
p_value_bar = TRUE,
colors = NULL,
x_lab = "description")
通路热图
data("metacyc_abundance")
data("metadata")
# Perform differential abundance analysis
metacyc_daa_results_df <- pathway_daa(
abundance = metacyc_abundance %>% column_to_rownames("pathway"),
metadata = metadata,
group = "Environment",
daa_method = "LinDA"
)
# Annotate the results
annotated_metacyc_daa_results_df <- pathway_annotation(
pathway = "MetaCyc",
daa_results_df = metacyc_daa_results_df,
ko_to_kegg = FALSE
)
feature_with_p_0.05 <- metacyc_daa_results_df %>%
filter(p_adjust < 0.05)
pathway_heatmap(
abundance = metacyc_abundance %>%
right_join(
annotated_metacyc_daa_results_df %>% select(all_of(c("feature","description"))),
by = c("pathway" = "feature")
) %>%
filter(pathway %in% feature_with_p_0.05$feature) %>%
select(-"pathway") %>%
column_to_rownames("description"),
metadata = metadata,
group = "Environment"
)

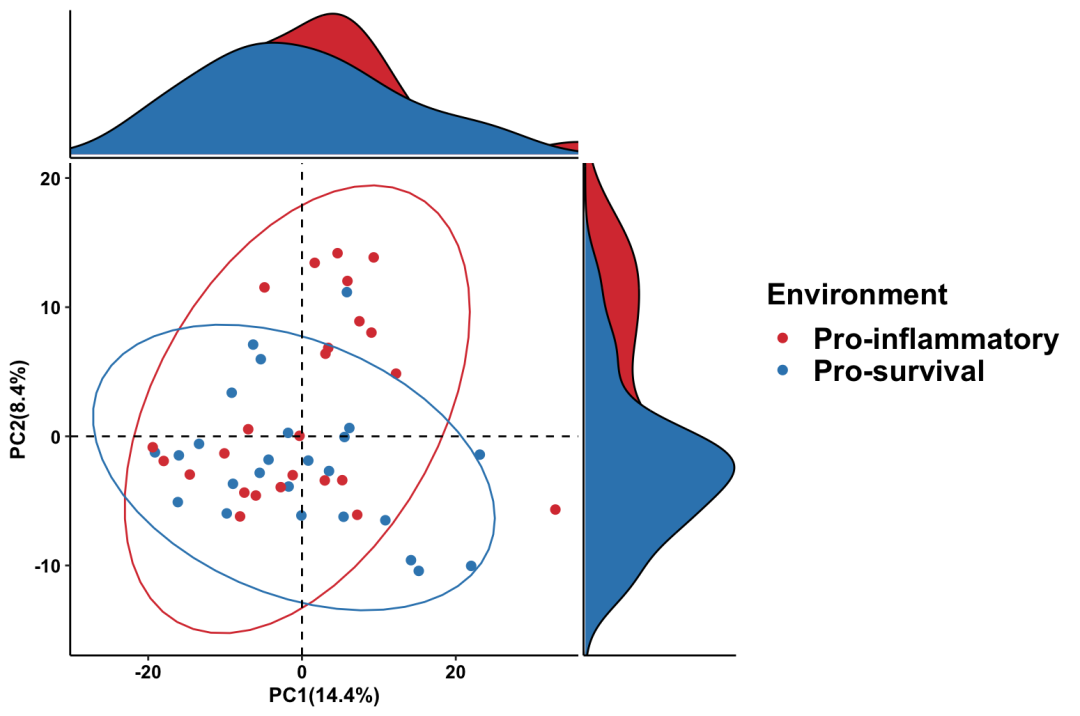
Pathway_PCA
pathway_pca(abundance = metacyc_abundance %>% column_to_rownames("pathway"),
metadata = metadata, group = "Environment")
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2023-08-19,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

