手把手带你复现NC之Figure7
复现文章信息:
文章题目:Single-cell analysis reveals prognostic fibroblast subpopulations linked to molecular and immunological subtypes of lung cancer 期刊:Nature Communications 日期:2023年1月31日 DOI: 10.1038/s41467-023-35832-6
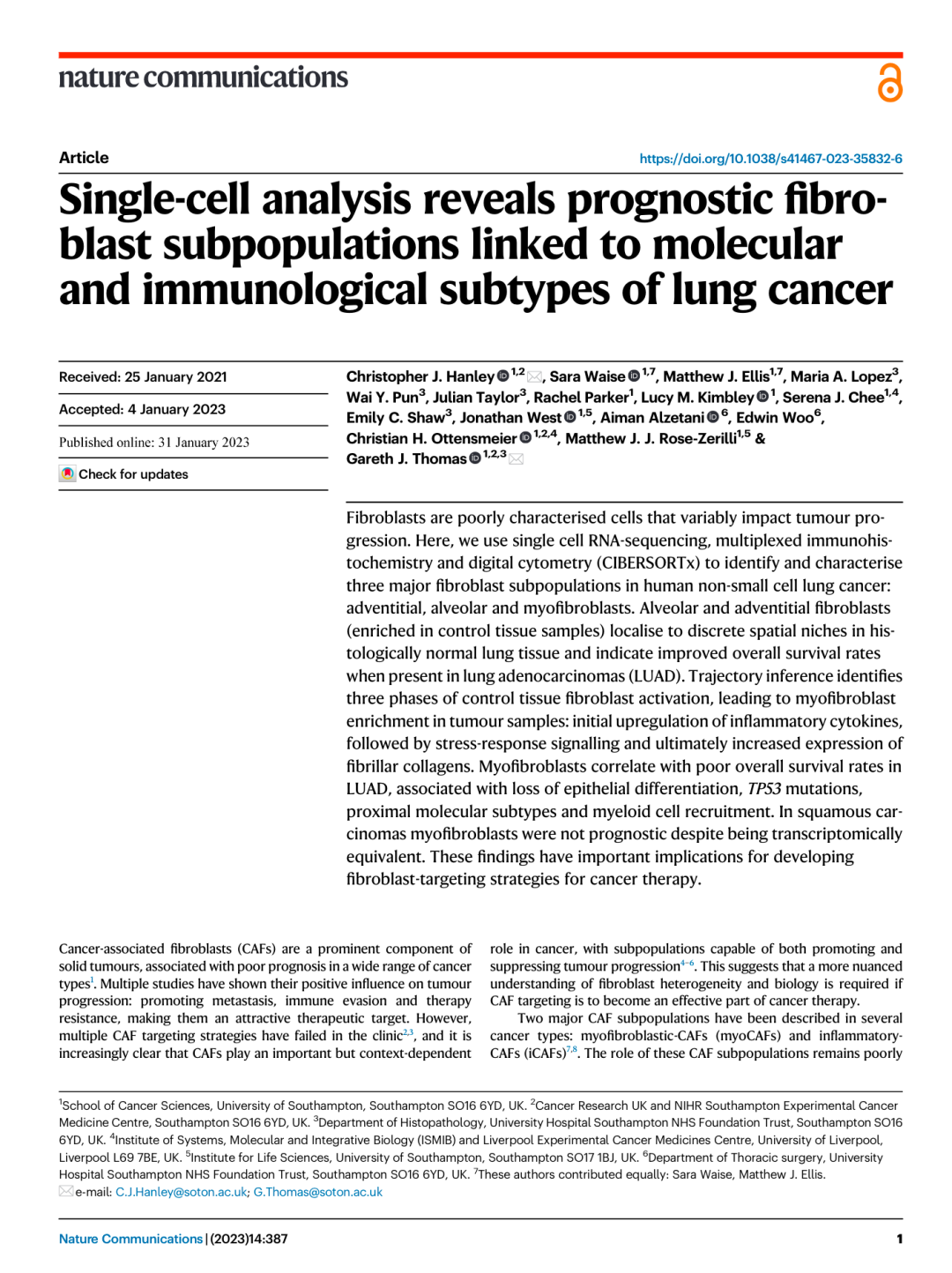
复现图——Figure7
显示成纤维细胞亚群与LUAD肿瘤的形态学、分子和免疫学特征关系
R包载入与数据准备
options(stringsAsFactors = F)
pardefault <- par()
library(Seurat)
library(tidyverse)
library(ggpubr)
library(ggsci)
library(survival)
library(survminer)
library(forestmodel)
data_directory <- "H:\\文献复现\\6\\"
source(paste0(data_directory, "0_NewFunctions.R"))
load(paste0(data_directory, "BulkData_Zenodo.Rdata"))
load(paste0(data_directory, "MxIHC_Zenodo.Rdata"))
load(paste0(data_directory, "IntegratedFibs_Zenodo.Rdata"))
LM22_names <- names(Merged.LUADtraits)[32:53]
fibs_names <- c("Adventitial_Fibs.pct", "Alveolar_Fibs.pct", "Myo_Fibs.pct")
Figure 7A
#G1 example
Slide = "Drop-Seq Cohort 8"
sample_data <- MxIHC_Fibroblasts %>% filter(Sample == Slide)
XY_cols <- names(sample_data)[1:2]
convex.hull.idx <- chull(as.matrix(sample_data[sample_data$Sample.Region.TvN == "Tumour", XY_cols]))
convex.hull.pts <- rownames(sample_data[sample_data$Sample.Region.TvN == "Tumour",])[convex.hull.idx]
G1_eg <-
sample_data %>%
slice_sample(prop = 0.25) %>%
ggplot(aes(x = X.Center..Pxl., y = Y.Center..Pxl., colour = Cell.type)) +
geom_point(size = 0.1) +
theme_void(base_size = 7) +
guides(colour = guide_legend(override.aes = list(size = 2))) +
theme(legend.position = "none") +
scale_color_manual(values = Fibs_col.palette) +
geom_polygon(data = MxIHC_Fibroblasts[convex.hull.pts, XY_cols],
colour = "black", alpha = 0, linetype = "dotted", size = 2)
G1_eg
#G2 example
Slide = "Lung Adeno 2"
sample_data <- MxIHC_Fibroblasts %>% filter(Sample == Slide)
XY_cols <- names(sample_data)[1:2]
convex.hull.idx <- chull(as.matrix(sample_data[sample_data$Sample.Region.TvN == "Tumour", XY_cols]))
convex.hull.pts <- rownames(sample_data[sample_data$Sample.Region.TvN == "Tumour",])[convex.hull.idx]
G2_eg <-
sample_data %>%
slice_sample(prop = 0.25) %>%
ggplot(aes(x = X.Center..Pxl., y = Y.Center..Pxl., colour = Cell.type)) +
geom_point(size = 0.1) +
theme_void(base_size = 7) +
guides(colour = guide_legend(override.aes = list(size = 2))) +
theme(legend.position = "none") +
scale_color_manual(values = Fibs_col.palette) +
geom_polygon(data = MxIHC_Fibroblasts[convex.hull.pts, XY_cols],
colour = "black", alpha = 0, linetype = "dotted", size = 2)
G2_eg
#G3 example
Slide = "Lung Adeno 5"
sample_data <- MxIHC_Fibroblasts %>% filter(Sample == Slide)
XY_cols <- names(sample_data)[1:2]
convex.hull.idx <- chull(as.matrix(sample_data[sample_data$Sample.Region.TvN == "Tumour", XY_cols]))
convex.hull.pts <- rownames(sample_data[sample_data$Sample.Region.TvN == "Tumour",])[convex.hull.idx]
G3_eg <-
sample_data %>%
slice_sample(prop = 0.25) %>%
ggplot(aes(x = X.Center..Pxl., y = Y.Center..Pxl., colour = Cell.type)) +
geom_point(size = 0.1) +
theme_void(base_size = 7) +
guides(colour = guide_legend(override.aes = list(size = 2))) +
theme(legend.position = "none") +
scale_color_manual(values = Fibs_col.palette) +
geom_polygon(data = MxIHC_Fibroblasts[convex.hull.pts, XY_cols],
colour = "black", alpha = 0, linetype = "dotted", size = 2)
G3_eg
which.max(c(max(layer_scales(G1_eg)$x$range$range),
max(layer_scales(G2_eg)$x$range$range),
max(layer_scales(G3_eg)$x$range$range)))
which.max(c(max(layer_scales(G1_eg)$y$range$range),
max(layer_scales(G2_eg)$y$range$range),
max(layer_scales(G3_eg)$y$range$range)))
xmax <- max(layer_scales(G1_eg)$x$range$range)
ymax <- max(layer_scales(G2_eg)$y$range$range)
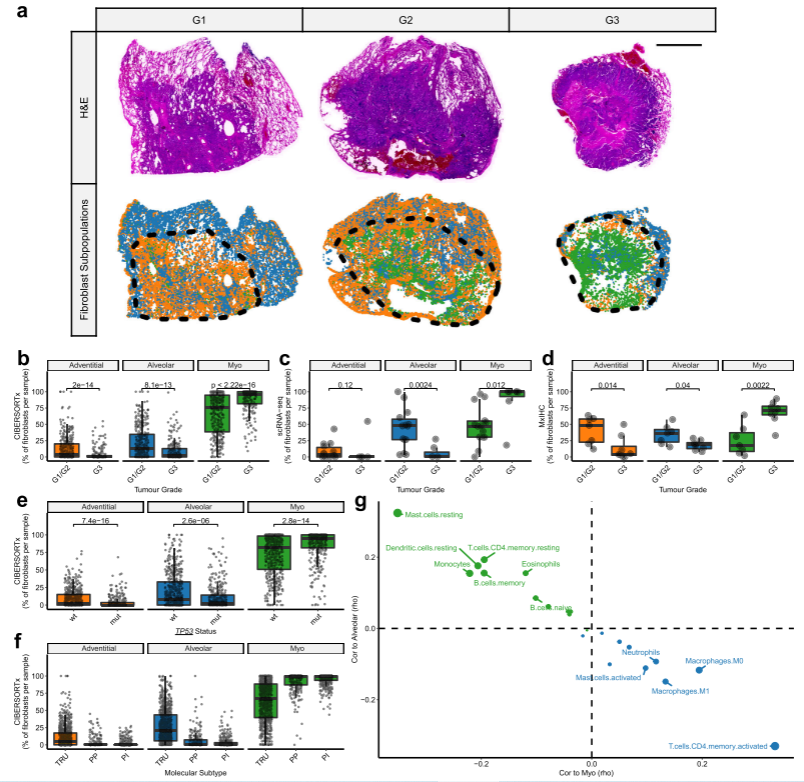
#来自不同级别LUAD肿瘤全切片mxIHC分析的代表性图像(G1 =高分化[n=1],G2 =中等分化[n=6]和G3 =低分化[n=8]),显示点模式图代表不同成纤维细胞亚群的空间分布(通过mxIHC数据的组织细胞分析测量)。黑色虚线划分每个组织切片中的肿瘤区域。
Fig_7A <-
ggarrange(G1_eg + expand_limits(y = c(median(layer_scales(G1_eg)$y$range$range) - 0.5*ymax,
median(layer_scales(G1_eg)$y$range$range) + 0.5*ymax)),
G2_eg + expand_limits(x = c(median(layer_scales(G2_eg)$x$range$range) - 0.5*xmax,
median(layer_scales(G2_eg)$x$range$range) + 0.5*xmax)),
G3_eg + expand_limits(x = c(median(layer_scales(G3_eg)$x$range$range) - 0.5*xmax,
median(layer_scales(G3_eg)$x$range$range) + 0.5*xmax),
y = c(median(layer_scales(G3_eg)$y$range$range) - 0.5*ymax,
median(layer_scales(G3_eg)$y$range$range) + 0.5*ymax)),
nrow = 1, ncol = 3)

Fig_7A
Figure 7B
箱形图显示高/中分化LUAD肿瘤与低分化LUAD肿瘤成纤维细胞亚群的相对丰度,通过CIBERSORTx测量
Bulk_Fibs.pct <- reshape2::melt(Merged.LUADtraits, id.vars = c("Grade2", "Dataset.factor", "Stage", "Stage_4cat", "TP53.Status", "EGFR.Status", "KRAS.Status","LUAD.MolSubtype_factor"),
measure.vars = c("Myo_Fibs.pct", "Alveolar_Fibs.pct", "Adventitial_Fibs.pct")
)
Bulk_Fibs.pct$Fibs_SubPop <- factor(
Bulk_Fibs.pct$variable,
levels = levels(Bulk_Fibs.pct$variable)[c(3,2,1)],
labels = c("Adventitial", "Alveolar", "Myo")
)
names(Bulk_Fibs.pct)[names(Bulk_Fibs.pct) == "value"] <- "Fibs.pct"
my.comparisons <- list(c("G1/G2", "G3"))
Fig_7B <-
Bulk_Fibs.pct %>%
drop_na(Grade2) %>%
ggplot(aes(x = Grade2, y = Fibs.pct)) +
theme_pubr(base_size = 15) +
facet_wrap(~Fibs_SubPop) +
theme(legend.position = "none") +
geom_boxplot(outlier.shape = NA, aes(fill = Fibs_SubPop)) +
geom_jitter(alpha = 0.5, width = 0.2, size = 0.1) +
scale_fill_manual(values = Fibs_col.palette) +
rotate_x_text(angle = 45) +
scale_y_continuous(limits = c(0,125), breaks = c(0,25,50,75,100)) +
stat_compare_means(comparisons = my.comparisons, size = 4) +
xlab("Tumour Grade") +
ylab("% of fibroblasts per sample\n(CIBERSORTx)")
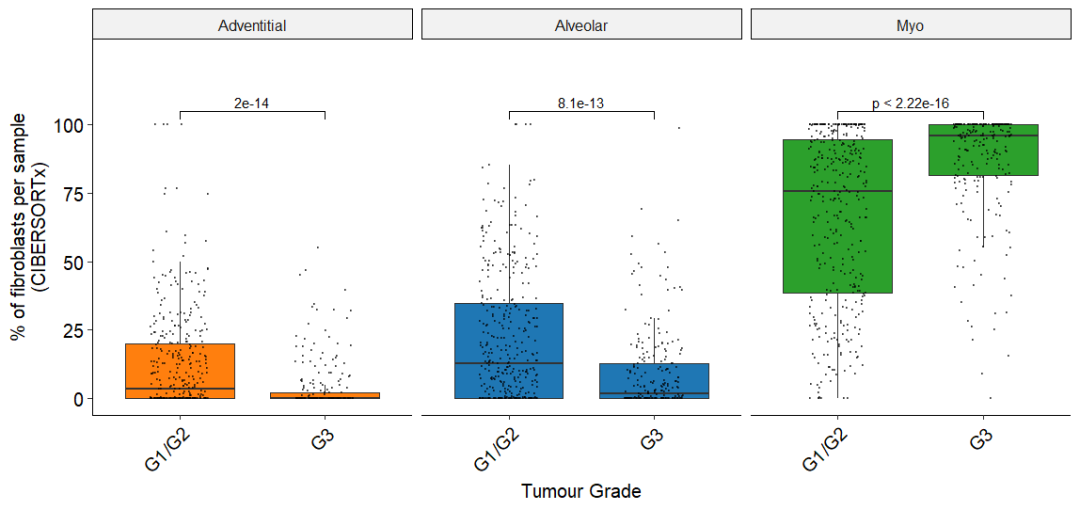
Fig_7B
LUAD肿瘤的形态学亚型被认为与患者生存率相关。如CIBERSORTx所示,LUAD形态学亚型间成纤维细胞亚群丰度存在显著差异
Figure 7C
箱形图显示高/中分化LUAD肿瘤与低分化LUAD肿瘤成纤维细胞亚群的相对丰度,通过scRNA-seq测定
Fibs.integrated$SampleID2 <- paste(Fibs.integrated$Dataset, Fibs.integrated$SampleID, sep = "_")
Sample_MetaData$SampleID2 <- paste(Sample_MetaData$Dataset, Sample_MetaData$SampleID, sep = "_")
Sample_MetaData$Sample.type[Sample_MetaData$Sample.type == "Tumor"] <- "Tumour"
Sample_MetaData$Sample.Subtype2 <- droplevels(Sample_MetaData$Sample.Subtype, exclude = c("Carcinoid" , "Pleiomorphic.carcinoma", "LCLC") )
dt <- as.table(as.matrix(table(Fibs.integrated$SampleID2,
Fibs.integrated$Fibs_MajorClusters)))
Sample.pct_long <- as.data.frame(dt/rowSums(dt)*100)
scRNAseq_Fibs.pct <- merge(Sample_MetaData, Sample.pct_long,
by.x = "SampleID2", by.y = "Var1")
names(scRNAseq_Fibs.pct)[23] <- "Fibs.pct"
names(scRNAseq_Fibs.pct)[22] <- "Fibs_SubPop"
scRNAseq_Fibs.pct$Fibs_SubPop <- factor(
scRNAseq_Fibs.pct$Fibs_SubPop,
levels = c("Adventitial", "Alveolar", "Myo")
)
scRNAseq_Fibs.pct$Differentiation2 <- factor(scRNAseq_Fibs.pct$Differentiation,
levels = c("G1", "G2", "G3"),
labels = c("G1/G2", "G1/G2", "G3"))
my.comparisons <- list(c("G1/G2", "G3"))
Fig_7C <-
scRNAseq_Fibs.pct %>%
filter(Sample.Subtype2 == "LUAD") %>%
drop_na(Differentiation2) %>%
ggplot(aes(x = Differentiation2, y = Fibs.pct)) +
theme_pubr(base_size = 15) +
theme(legend.position = "none") +
facet_wrap(~Fibs_SubPop) +
geom_boxplot(outlier.shape = NA, aes(fill = Fibs_SubPop)) +
geom_jitter(alpha = 0.5, width = 0.2) +
scale_fill_manual(values = Fibs_col.palette) +
rotate_x_text(angle = 45) +
scale_y_continuous(limits = c(0,125), breaks = c(0,25,50,75,100)) +
stat_compare_means(comparisons = my.comparisons, size = 5) +
xlab("Tumour Grade") +
ylab("% of fibroblasts per sample\n(scRNA-seq)")

Fig_7C
Figure 7D
箱形图显示高/中分化LUAD肿瘤与低分化LUAD肿瘤成纤维细胞亚群的相对丰度,通过mxIHC测定
MxIHC_Fibroblasts$TvN.Grouping.ID <- paste(MxIHC_Fibroblasts$Slide.name, MxIHC_Fibroblasts$Sample.Region.TvN, sep = "_")
dt <- as.table(as.matrix(table(MxIHC_Fibroblasts$TvN.Grouping.ID,
MxIHC_Fibroblasts$Cell.type)))
Sample.pct_long <- as.data.frame(dt/rowSums(dt)*100)
Sample.pct_long$Slide.name <- do.call(rbind, strsplit(as.character(Sample.pct_long$Var1), "_", fixed = T))[,1]
Sample.pct_long$TvN <- do.call(rbind, strsplit(as.character(Sample.pct_long$Var1), "_", fixed = T))[,2]
TvN.Data_long <- merge(MxIHC_Sample.metaData, Sample.pct_long,
by = "Slide.name")
names(TvN.Data_long)[names(TvN.Data_long) == "Freq"] <- "Fibs.pct"
names(TvN.Data_long)[names(TvN.Data_long) == "Var2"] <- "Fibs_SubPop"
TvN.Data_long$SubtypeTvN <- TvN.Data_long$Subtype
TvN.Data_long$SubtypeTvN[TvN.Data_long$TvN == "Control"] <- "Control"
MxIHC_Fibs.pct <- TvN.Data_long[!TvN.Data_long$TvN == "NA", ]
names(MxIHC_Fibs.pct)
MxIHC_Fibs.pct$Differentiation2 <- factor(MxIHC_Fibs.pct$Grade,
levels = c("G1", "G2", "G3"),
labels = c("G1/G2", "G1/G2", "G3"))
Fig_7D <-
MxIHC_Fibs.pct %>%
filter(SubtypeTvN == "LUAD") %>%
drop_na(Differentiation2) %>%
ggplot(aes(x = Differentiation2, y = Fibs.pct)) +
theme_pubr(base_size = 15) +
theme(legend.position = "none") +
facet_wrap(~Fibs_SubPop) +
geom_boxplot(outlier.shape = NA, aes(fill = Fibs_SubPop)) +
geom_jitter(alpha = 0.5, width = 0.2) +
scale_fill_manual(values = Fibs_col.palette) +
rotate_x_text(angle = 45) +
scale_y_continuous(limits = c(0,125), breaks = c(0,25,50,75,100)) +
stat_compare_means(comparisons = my.comparisons, size = 4, label.y = 100) +
xlab("Tumour Grade") +
ylab("% of fibroblasts per sample\n(MxIHC)")
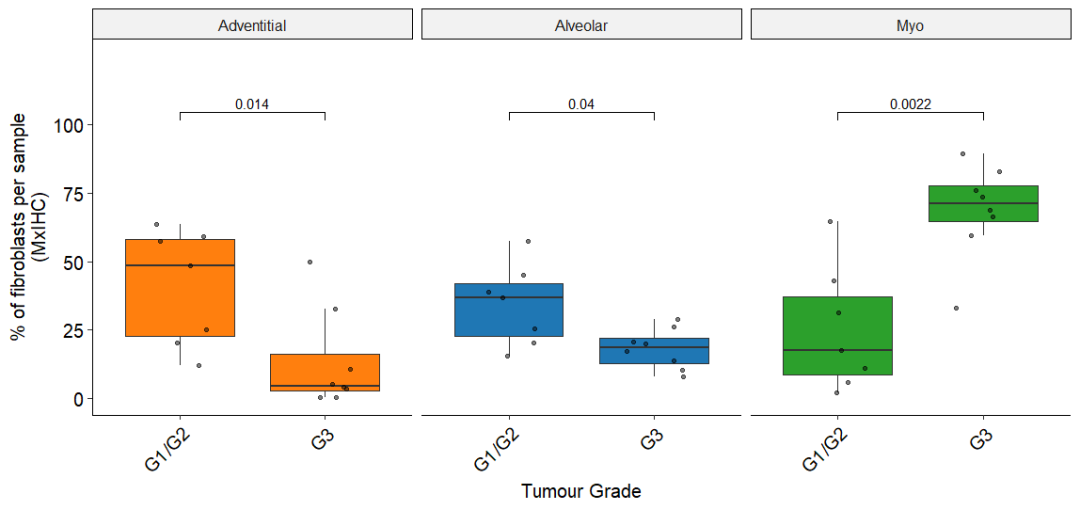
Fig_7D
Figure 7E
箱线图显示了通过CIBERSORTx测量的按TP53突变状态分组的LUAD肿瘤中成纤维细胞亚群的相对丰度
Fig_7E <-
Bulk_Fibs.pct %>%
drop_na(TP53.Status) %>%
ggplot(aes(x = TP53.Status, y = Fibs.pct)) +
theme_pubr(base_size =15) +
theme(legend.position = "none") +
facet_grid(~Fibs_SubPop, scales = "free_y") +
geom_boxplot(outlier.shape = NA, aes(fill = Fibs_SubPop)) +
geom_jitter(alpha = 0.5, width = 0.2, size = 0.1) +
scale_fill_manual(values = Fibs_col.palette) +
rotate_x_text(angle = 45) +
scale_y_continuous(limits = c(0,125), breaks = c(0,25,50,75,100)) +
stat_compare_means(comparisons = list(c("0","1")), size = 4,
label.y = 110) +
xlab("TP53 Status") +
scale_x_discrete(labels = c("wt", "mut")) +
ylab("% of fibroblasts per sample\n(CIBERSORTx)")

Fig_7E
在含有TP53突变的LUAD肿瘤中,肌成纤维细胞丰度增加
Figure 7F
箱形图显示按分子亚型(TRU终末呼吸单位、PP近端增生性和PI近端炎症)分组的LUAD肿瘤中成纤维细胞亚群的相对丰度。每个数据点代表通过CIBERSORTx测量的个体患者
Fig_7F <-
Bulk_Fibs.pct %>%
drop_na(LUAD.MolSubtype_factor) %>%
ggplot(aes(x = LUAD.MolSubtype_factor, y = Fibs.pct)) +
theme_pubr(base_size = 15) +
theme(legend.position = "none") +
facet_grid(~Fibs_SubPop, scales = "free_y") +
geom_boxplot(outlier.shape = NA, aes(fill = Fibs_SubPop)) +
geom_jitter(alpha = 0.5, width = 0.2, size = 0.1) +
scale_fill_manual(values = Fibs_col.palette) +
rotate_x_text(angle = 45) +
scale_y_continuous(limits = c(0,125), breaks = c(0,25,50,75,100)) +
xlab("Molecular Subtype") +
scale_x_discrete(labels = c("TRU", "PP", "PI")) +
ylab("% of fibroblasts per sample\n(CIBERSORTx)")
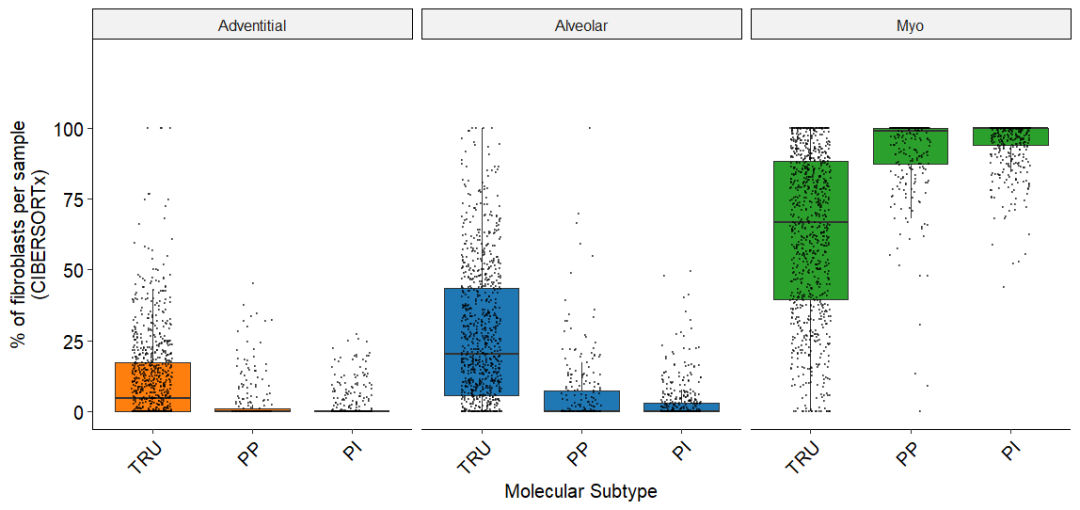
Fig_7F
在大量组织数据集中发现77%的TRU肿瘤为中度/高分化(G1/G2), 69%的PP肿瘤为低分化(G3)。正如预期的那样,考虑到这种联系,肌成纤维细胞丰度在PP肿瘤中最高;而肺泡和表皮成纤维细胞在TRU肿瘤中最为突出
Figure 7G
散点图显示肺泡或肌成纤维细胞丰度与免疫细胞亚群(LM22)丰度之间的Spearman相关性,通过CIBERSORTx测量
LM22_names <- names(Merged.LUADtraits)[32:53]
fibs_names <- c("Adventitial_Fibs.pct", "Alveolar_Fibs.pct", "Myo_Fibs.pct")
Immune_cor <- WGCNA::corAndPvalue(Merged.LUADtraits[, LM22_names],
Merged.LUADtraits[, fibs_names],
use = "pairwise.complete.obs",
method = "spearman")
Immune_cor$sig_cor <- Immune_cor$cor
Immune_cor$sig_cor[Immune_cor$p > 0.001] <- NA
Immune_cor$cor_df <- data.frame(
Immune_cor$cor,
Immune.cell = rownames(Immune_cor$cor)
)
Immune_cor$cor_df$Immune.cell.label <- Immune_cor$cor_df$Immune.cell
sig_cors <- rownames(na.omit(Immune_cor$sig_cor[,2:3]))
Immune_cor$cor_df$Immune.cell.label[!Immune_cor$cor_df$Immune.cell %in% sig_cors] <- NA
Fig_7G <-
Immune_cor$cor_df %>%
ggplot(aes(y = Alveolar_Fibs.pct, x = Myo_Fibs.pct,
colour = Alveolar_Fibs.pct > Myo_Fibs.pct)) +
theme_pubr(base_size = 15) +
geom_hline(yintercept = 0, linetype = "dashed") +
geom_vline(xintercept = 0, linetype = "dashed") +
geom_point(aes(size = abs(Myo_Fibs.pct))) +
scale_size(range = c(0.1, 2)) +
theme(legend.position = "none") +
scale_color_manual(values = c(Fibs_col.palette[[2]],Fibs_col.palette[[3]])) +
ggrepel::geom_text_repel(aes(label = Immune.cell.label),
size = 4, min.segment.length = 0) +
xlab("Cor to Myo (rho)") +
ylab("Cor to Alveolar (rho)")
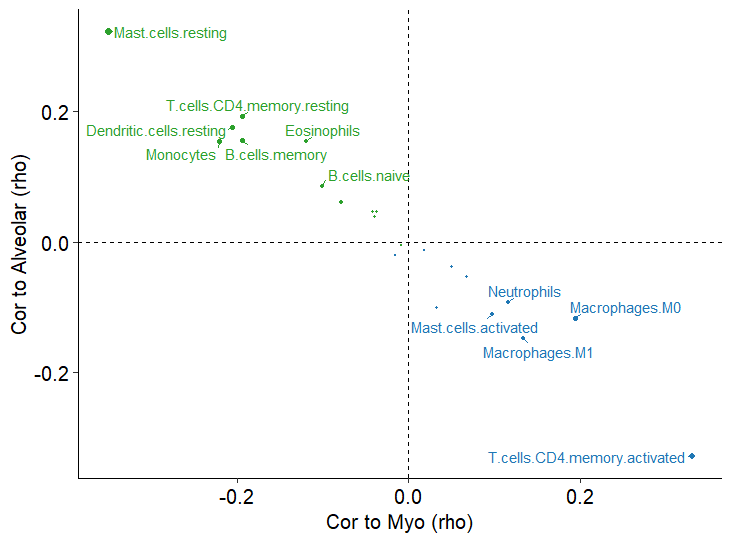
Fig_7G
使用CIBERSORTx检测免疫细胞亚群(LM22)的丰度及其与成纤维细胞亚群的相关性。表明与肌成纤维细胞和肺泡成纤维细胞相关的免疫细胞之间存在反比关系,这在分析的所有LUAD转录组数据集中一致地观察到。肺泡成纤维细胞与多个静息免疫细胞亚群相关。相反,肌成纤维细胞与巨噬细胞、中性粒细胞、活化肥大细胞和活化CD4 +记忆T细胞相关。表明肿瘤微环境中的肌成纤维细胞分化也与辅助性T细胞和骨髓细胞的激活/分化有关。
小结
- 本项研究确定了存在于对照组织和肿瘤组织中的三个主要亚群:外膜成纤维细胞、肺泡成纤维细胞和肌成纤维细胞。还表明了癌症相关的肌成纤维细胞可能起源于组织内的外基质或肺泡成纤维细胞,这一过程涉及炎症和应激反应信号的短暂阶段。成纤维细胞亚群丰度在对照组织和肿瘤组织之间以及在非小细胞肺癌组织学亚型之间存在差异,在LUAD中具有重要的预后价值,因为它们与这些肿瘤的分子、形态学和免疫特征有关。
- 在对照组织样本中常见的两种成纤维细胞亚群(肺泡和外膜成纤维细胞)与LUAD患者生存率的提高有关,尤其是肺泡成纤维细胞。在跨组织分析表明,肺泡成纤维细胞对肺组织具有相对特异性(与外膜和肌成纤维细胞群体相比)。然而,这并不能排除在其他器官中存在类似的肿瘤抑制的常驻成纤维细胞群。
- 外膜成纤维细胞亚群与先前的多项研究一致,可能代表普遍的成纤维细胞。研究发现CD34是该群体的有效标记物,这与许多描述不同器官中CD34+ 成纤维细胞/间质细胞群体的研究一致。然而与之前的研究相反,研究发现炎症相关细胞因子和iCAF标记物的表达主要局限于非小细胞肺癌的对照组织。此外,轨迹分析表明,这种炎症类型可能代表了向肌成纤维细胞表型转移的早期和短暂阶段,独立于与肿瘤的直接相互作用。事实上,炎性成纤维细胞已经在感染、炎症条件和癌症中被发现,这支持了成纤维细胞通过一系列表型发展的概念,这些表型的功能是启动、支持和最终抑制炎症。
- 研究结果表明,这些不一致可能是由于成纤维细胞的异质性和非小细胞肺癌组织学亚型的差异。这可能是因为LUSC(与LUAD相比)具有更均匀的基质,含有持续高水平的肌成纤维细胞,这可能限制了其作为预后标志物的效用。另外,肌成纤维细胞对肿瘤进展的功能影响可能因肿瘤亚型而异。这些细胞如何促进肿瘤进展的多种机制已经在临床前癌症模型中被确定。然而,最近的研究也表明,肌成纤维细胞在PDAC中具有肿瘤抑制特性。观察到肌成纤维细胞对LUSC的预后没有影响,这进一步证明了这些细胞在癌症中所起的作用可能与环境有关,需要进一步的研究来确定这种差异背后的机制。鉴于我们发现来自LUAD和LUSC肿瘤的肌成纤维细胞之间的基因表达没有显著差异,这些细胞对肿瘤进展的不同影响可能是由于恶性细胞的内在特性,正如最近一项研究所描述的那样,阐明了LUAD对GREM1-KDR信号在疾病进展中的特异性作用
- 总之,这项对肺和非小细胞肺癌成纤维细胞异质性的综合分析显示,某些亚群是器官特异性的,而其他亚群则在不同的组织、癌症和病理中一致存在。肌成纤维细胞和肺泡成纤维细胞与不同的LUAD分子和免疫亚型相关,它们对预后的相反作用准确地识别了高危患者。这些发现可以改善患者分层,并应该完善肺癌成纤维细胞的治疗靶向策略。