R和python绘制柱状堆叠图技巧
R和python绘制柱状堆叠图技巧
生信技能树jimmy
发布于 2024-03-07 10:10:23
发布于 2024-03-07 10:10:23
代码可运行
文章被收录于专栏:单细胞天地
运行总次数:0
代码可运行
柱状堆叠图在许多的单细胞文章中被使用,通过它可以更好向我们展示一个亚群中各个细胞的占比情况,可见它如此重要!!!今天,我们来使用R的ggplot2和python的matplotlib分别绘制柱状堆叠图。
绘制技巧如下图所示:
先画出图的大致轮廓,再根据需求,添加更多的细节和细节调整,一张完美的图就出来了啊!
R的ggplot2绘图
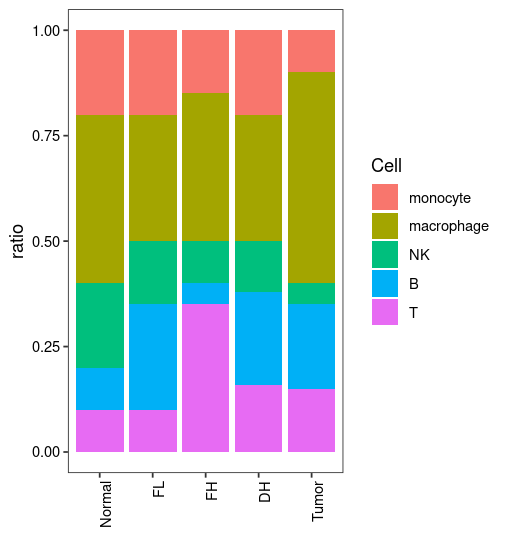
这是一张5个样本中各种免疫细胞比例情况的可视化
1.模拟数据:
library(reshape2)
library(ggplot2)
#模拟数据并创建一个5样本细胞数据集
status=c('Normal','FL','FH','DH','Tumor')
monocyte=c(0.2,0.2,0.15,0.2,0.1)
macrophage=c(0.4,0.3,0.35,0.3,0.5)
NK=c(0.2,0.15,0.1,0.12,0.05)
B=c(0.1,0.25,0.05,0.22,0.2)
T=c(0.1,0.1,0.35,0.16,0.15)
#创建数据框
data=data.frame(status=status,monocyte=monocyte,macrophage=macrophage,NK=NK,B=B,T=T)
data$Status <- factor(data$status, c('Normal','FL','FH','DH','Tumor'))
#ggplot2画图需要宽数据变成长数据
melt.data <- melt(data, variable.name = 'Cell', value.name = 'ratio')
#配色
colors=c('#F08784','#A3A500','#00BF7D','#00B0F6','#E76BF3')
2.绘制柱状堆叠图的大致轮廓
p=ggplot(melt.data ,aes(x = Status, y = ratio, fill = Cell)) +
geom_bar(stat="identity")
3.添加细节和修改
p=p+scale_fill_manual(values =colors ) + #添加柱状堆叠图颜色
theme_bw() +
theme(axis.text = element_text(colour = 'black'),
axis.text.x = element_text(angle = 90, hjust = 1),
panel.grid = element_blank()) + #设置主题背景
labs(x = '') #修改x轴的标题
Python的matplotlib绘图
#安装matplotlib、numpy和pandas库,如果之前有安装,就不用安装了
#在pycharm终端输入下面命令就可以进行安装了
pip install matplotlib numpy pandas -i http://pypi.tuna.tsinghua.edu.cn/simple
1.模拟数据:
#导入相关可视化库
import matplotlib.pyplot as plt,numpy as np,pandas as pd
#模拟数据并创建一个5样本细胞数据集
status=['Normal','FL','FH','DH','Tumor']
group=['T','B','NK','macrophage','monocyte']
data={'monocyte':[0.2,0.2,0.15,0.2,0.1],
'macrophage':[0.4,0.3,0.35,0.3,0.5],
'NK':[0.2,0.15,0.1,0.12,0.05],
'B':[0.1,0.25,0.05,0.22,0.2],
'T':[0.1,0.1,0.35,0.16,0.15]
}
#与R类似相当于创建一个数据框
data=pd.DataFrame(data)
#配色
colors=['#E76BF3','#00B0F6','#00BF7D','#A3A500','#F08784']
2.绘制柱状堆叠图的大致轮廓
bottom = np.zeros(5)
fig, axes = plt.subplots(figsize=(6,5),ncols=2,width_ratios=[5,1])
ax1, ax2 = axes.ravel()
ax2.axis('off')
p=[]
for k,i in enumerate(group):
patches=ax1.bar(status, data[i].tolist(), label=i,color=colors[k],bottom=bottom)
p.append(patches)
bottom += data[i].values
3.添加细节和修改
#修改Y轴的标题
ax1.set_ylabel('ratio')
#添加图例和其位置调整
ax2.legend(p, group, loc='center left',frameon=False,prop={'size':12},title='Cell')
plt.tight_layout()
plt.show()
可视化结果:

看起来是不是挺简单的,感兴趣的小伙伴可以进行细节微调,进一步美化图形,我们下次再见!
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2024-03-06,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
