空间转录组学数据分析细胞邻域依赖的基因表达(分子邻域)
原创空间转录组学数据分析细胞邻域依赖的基因表达(分子邻域)
原创
追风少年i
发布于 2024-03-25 15:49:22
发布于 2024-03-25 15:49:22
作者,Evil Genius
今天我们来继续分享关于空间转录组的内容,回到之前的一个分析,空间转录组有四大矩阵
1、分子矩阵,gene X barcode,这是最开始大家拿到的矩阵 2、细胞矩阵,空间解卷积之后的矩阵,细胞 X Barcode 3、分子niche矩阵, 即分子生态位矩阵,主要研究分子微环境,包括邻域通讯等, gene X Barcode 4、细胞niche矩阵,即细胞生态位矩阵,主要用来研究细胞的空间排布,例如侵袭性的肿瘤细胞空间临近巨噬,所有细胞的空间排布形成了细胞niche矩阵.
今天我们要分享关于第三个矩阵的分析,即分子niche矩阵,主要的目的就是研究细胞邻域依赖基因表达。
其中涉及到的内容,细胞邻域,细胞类型,基因表达。
- 细胞会根据其相邻细胞的不同类型表达不同的基因,这些基因与发育或转移等关键生物过程有关.
- 邻域依赖性基因表达表明,除了配体-受体共表达所能发现的基因外,还有新的潜在基因参与了细胞-细胞间的相互作用
细胞已经进化出它们的通讯方法来感知它们的微环境并发送生物信号。除了使用配体和受体进行通信外,细胞还使用包括间隙连接在内的多种通道与其近邻进行通信。邻接依赖性基因表达提示了在配体-受体共表达之外参与细胞-细胞相互作用的新的潜在基因。
cell–cell interactions; cellular communication; neighbor-dependent genes
细胞以多种方式与其微环境进行交流,包括释放可溶性分子和直接细胞接触,积极改变其转录组以响应外部信号。为了深入了解关键的生物过程,如疾病和发育,了解细胞间交流的各种方式是必不可少的。研究细胞间通讯的实验方法通常需要精心设计和复杂的设置。
利用配体-受体共表达的单细胞RNA测序(scRNA-seq)和空间转录组学(ST)数据,可以在基因组尺度上研究细胞-细胞相互作用。使用配体-受体共表达可以推断相互作用的细胞类型对并识别细胞间信号通路,而无需依赖于复杂的实验设置。然而,它不能解释细胞直接接触所改变的单个细胞的基因表达。
越来越多的关于细胞通讯的研究表明,细胞受其微环境和邻近细胞的影响。
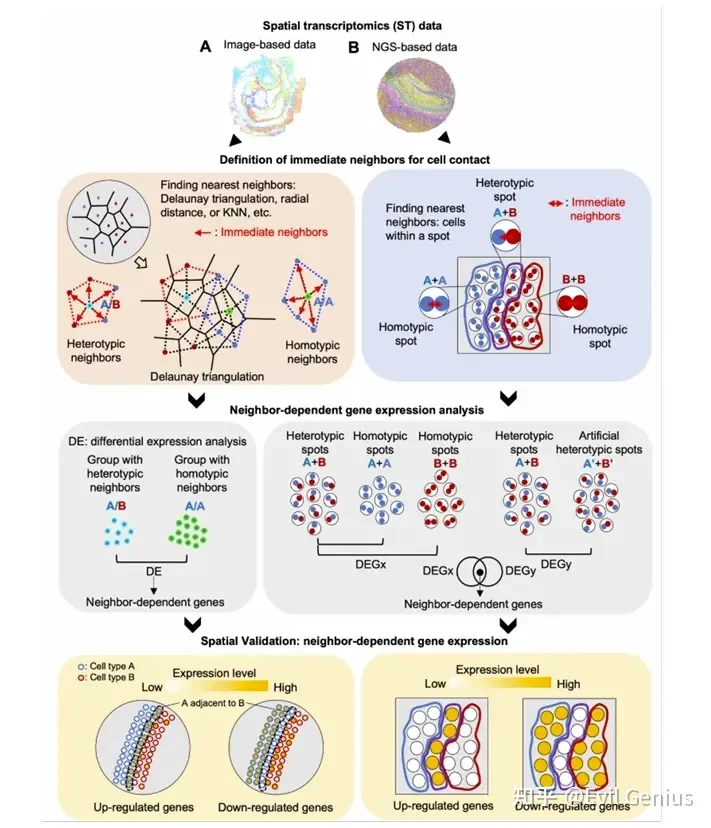
ST的最新发展为探索微环境的作用开辟了潜在的途径。空间基因表达谱使得研究细胞的转录活性以及完整组织内邻近细胞的转录活性成为可能。ST数据主要有两种类型,基于图像和基于NGS。
已经开发了许多计算工具来从ST数据中理解细胞-细胞相互作用。CellphoneDB v.3.0、MESSI、SpaTalk和stLearn使用配体-受体对的共表达来研究细胞通讯。然而,由于直接接触,配体-受体共表达不能完全捕获细胞-细胞相互作用。SVCA将基因表达变异的来源分解为内在效应、环境效应和细胞-细胞相互作用。它解释了基因表达和细胞间相互作用之间的关系。然而,SVCA不具有检测与细胞接触相关的基因表达变化的功能,并且它们的策略仅针对基于图像的ST数据进行了优化。由于MISTy量化了不同空间背景对感兴趣标记表达的贡献,因此可以研究近邻对标记表达的影响。然而,MISTy需要预先选择标记基因列表来发现潜在的相互作用,并且它还没有被设计成以一种公正的方式识别与细胞接触相关的基因表达变化。DeepLinc从ST数据重建细胞相互作用网络。将三个最近的邻居作为直接接触,DeepLinc发现了有助于细胞类型之间相互作用的特征基因,并推断出它们之间的近端相互作用。然而,它并没有揭示特征基因和相互作用的细胞类型之间的特定关系。C-SIDE检查上调和下调的基因取决于接近某种细胞类型。由于细胞类型之间的相互作用是基于细胞密度而不是细胞接触来定义的,因此C-SIDE不适用于研究细胞接触依赖性基因表达。NCEM研究依赖于局部环境的转录组变化,但它并没有被设计用于研究细胞接触对基因表达的影响,特别是对于基于NGS的数据。即使在低分辨率的Visium数据中,NCEM也将一个条形码点视为单个细胞类型,因此它不会研究一个点内多个细胞类型直接接触的影响。虽然空间环境已被应用于研究细胞-细胞相互作用,但与细胞接触相关的转录组学变化尚未得到充分探索。
邻域分子的分析策略
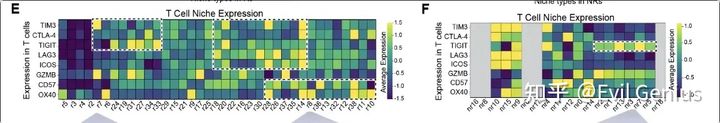
邻域依赖基因是参与细胞-细胞相互作用的一种新的潜在基因
邻居依赖基因表现出niche特异性表达
niche特异性基因表达解释了细胞异质性
我们来用代码分析一下这个问题,python版本,10X数据、华大数据、slide-seq数据都兼容,其他数据需要大家稍作修改
import pandas as pd
import os
import CellNeighborEX
# Check the path of your root directory.
os.getcwd()
# Make a folder to save data files.
if not os.path.exists('Datasets'):
os.makedirs('Datasets')
# Download data files.
# (i) pre-processed expression data
!wget https://figshare.com/ndownloader/files/42334083 -O Datasets/SSliver_log_data.txt
!wget https://figshare.com/ndownloader/files/42334077 -O Datasets/SSliver_cell_id.txt
!wget https://figshare.com/ndownloader/files/42334080 -O Datasets/SSliver_gene_name.txt
# (ii) data of annotated cell types and spatial coordinates
!wget https://figshare.com/ndownloader/files/42333705 -O Datasets/SSliver_RCTD.csv
##Load data
# Set the path of data files regarding annotated cell types.
path = '/Users/kimh15/Downloads/Datasets/'
df_processed = pd.read_csv(path + 'SSliver_RCTD.csv', header=0)生成按细胞类型分类的数据文件
# All categorzied files (index_, matchComb_, neiCombUnique_, prop_ .csv) are saved in the "categorized_data folder" in the root directory.
CellNeighborEX.categorization.generate_input_files(data_type = "NGS", df = df_processed, sample_size=30, min_sample_size=1)
# Set the path of the directory where all the categorized data files are saved.
path_categorization = '/Users/kimh15/Downloads/categorized_data/'
####Get log-normalized expression data
# Save the data into dataframes.
df_cell_id = pd.read_csv(path + "SSliver_cell_id.txt", delimiter="\t", header=None)
df_gene_name = pd.read_csv(path + "SSliver_gene_name.txt", delimiter="\t", header=None)
df_log_data = pd.read_csv(path + "SSliver_log_data.txt", delimiter="\t", header=None)邻域依赖基因表达分析,拿到每种细胞类型的空间依赖基因。
# Set argument values for CellNeighborEX.DEanalysis.analyze_data().
data_type = "NGS" # Image: image-based ST data, NGS: NGS-based ST data
lrCutoff = 0.4 # log ratio
pCutoff = 0.01 # p-value
pCutoff2 = 0.01 # false discovery rate
direction = 'up' # up: up-reguated genes, down: down-regulated genes
normality_test = False # True: depending on the result of the normality test, the statistical test is determined. If the data is normal, the parametric test is used. Otherwise, the non-parametric test is used.
# False: when sample size (number of cells/spots) is larger than 30, the parameteric test is used. Otherwise, the non-parametric test is used.
top_genes = 10 # Top 10 DEGs are annotated in the volcano plot.
# If save=True, all result files (DEG list: csv, heatmaps and volcano plots: pdf, gene expression values: txt) are saved in the "DE_results" folder in the root directory.
DEG_list = CellNeighborEX.DEanalysis.analyze_data(df_cell_id, df_gene_name, df_log_data, path_categorization, data_type, lrCutoff, pCutoff, pCutoff2, direction, normality_test, top_genes, save=True)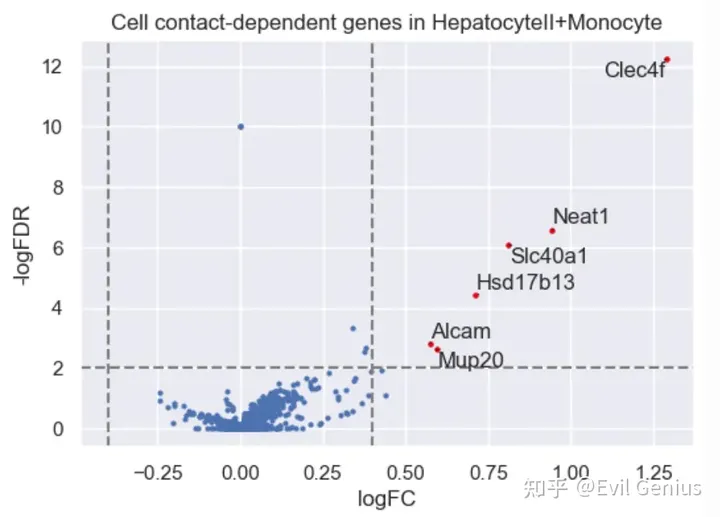
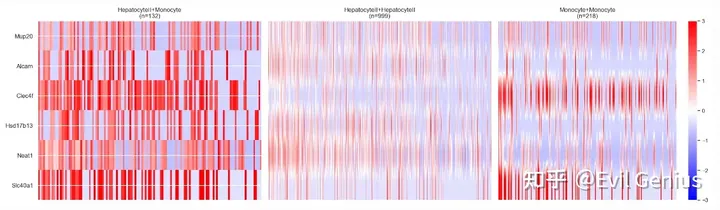
在空间数据中可视化邻居依赖的基因表达
# Select a cell type and a DEG for spatial visualization and then load the data.
# For example, F13a1 is one of up-regulated genes identified from the heterotypic spots of TumorIII+Monocyte.
path_selected = '/Users/kimh15/Downloads/DE_results/TumorIII+Monocyte/'
column_names = ['barcode', 'logdata', 'zscore']
heterotypic = pd.read_csv(path_selected + "TumorIII+Monocyte_F13a1.txt", delimiter=",", names = column_names)
homotypic1 = pd.read_csv(path_selected + "TumorIII+TumorIII_F13a1.txt", delimiter=",", names = column_names)
homotypic2 = pd.read_csv(path_selected + "Monocyte+Monocyte_F13a1.txt", delimiter=",", names = column_names)
heterotypic['type'] = 'TumorIII+Monocyte'
homotypic1['type'] = 'TumorIII+TumorIII'
homotypic2['type'] = 'Monocyte+Monocyte'
df_exp = pd.concat([heterotypic, homotypic1, homotypic2])
# Set parameter values.
df_bg, df_red, df_blue, df_black = CellNeighborEX.visualization.set_parameters(df_processed, df_exp, beadsize_bg=10, edgecolor_bg=(0.85,0.85,0.85), beadcolor_bg=(0.85,0.85,0.85), beadsize_red=600, beadsize_blue=200, beadsize_black=200, type_red='TumorIII+Monocyte', type_blue='TumorIII+TumorIII', type_black='Monocyte+Monocyte')
# Get the spatial map.
# zorder_red, zorder_blue, and zorder_black are parameters that determine the drawing order in the spatial map.
# If save=True, the spatial map (F13a1.pdf) is saved in the "spatialMap" folder in the root directory.
CellNeighborEX.visualization.get_spatialPlot(df_bg, df_red, df_blue, df_black, label_red='TumorIII+Monocyte', label_blue='TumorIII', label_black='Monocyte', label_gene='F13a1', zorder_red=3.0, zorder_blue=2.0, zorder_black=4.0, figsize=(28,28), save=True)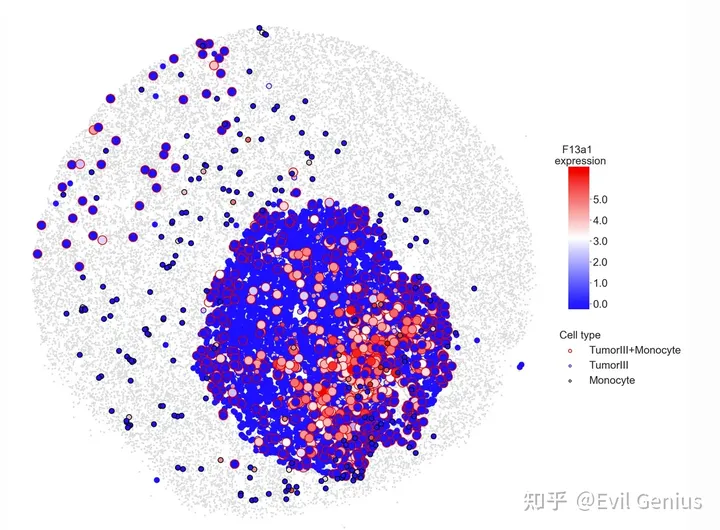
参考文献CellNeighborEX: deciphering neighbor‐dependent gene expression from spatial transcriptomics data
参考网址CellNeighborEX
生活很好,有你更好
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
原创声明:本文系作者授权腾讯云开发者社区发表,未经许可,不得转载。
如有侵权,请联系 cloudcommunity@tencent.com 删除。
评论
登录后参与评论
推荐阅读
目录
相关产品与服务
腾讯云代码分析
腾讯云代码分析(内部代号CodeDog)是集众多代码分析工具的云原生、分布式、高性能的代码综合分析跟踪管理平台,其主要功能是持续跟踪分析代码,观测项目代码质量,助力维护团队卓越代码文化。
