多组cellchat细胞通讯批量分析
上面是之前我们分享的数据集处理过程,今天我们来看看如何进行cellchat分析:1.批量运行多组cellchat 2.比较两组cellchat结果
今天是完整代码分享,如果想要使用其他示例数据,可以参考这个数据:单细胞直播一理解seurat数据结构与pbmc处理流程
下面进入正题:
数据准备1
#request 2
.libPaths(c( "/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2",
"/home/data/t040413/R/yll/usr/local/lib/R/site-library",
"/refdir/Rlib/", "/usr/local/lib/R/library"))
getwd()
filepath="~/biye/ipf/gse104154/cellchat"
dir.create(filepath,recursive = TRUE)
setwd(filepath)
getwd()
library(Seurat)
library(CellChat)
#####1 macrophage----
load("/home/data/t040413/ipf/GSE104154_scRNA-seq_fibrotic MC_bleomycin/allmerge.rds")
#load("~/silicosis/silicosis_cluster_merge.rds")
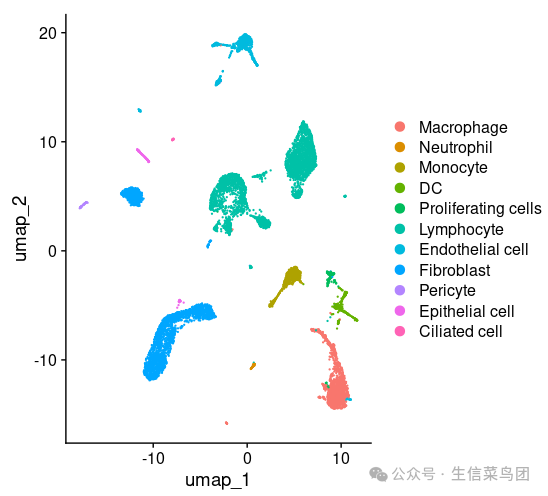
All.merge$cell.type=Idents(All.merge)
table(All.merge$stim)
All.merge$group=ifelse(grepl(pattern = '1|2|3',x = All.merge$stim),'Control','Bleomycin')
table(All.merge$group)
DimPlot(All.merge)
数据准备2
library(Seurat)
library(ggplot2)
subset_data=All.merge
subset_data$cell_type=Idents(subset_data)
table(subset_data$cell.type)
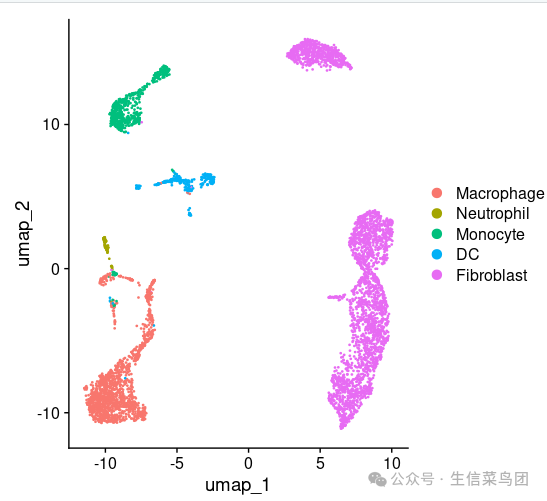
subset_data=subset(subset_data,idents= c('Macrophage',
'Neutrophil', 'Monocyte' , 'DC','Fibroblast'))
ggplot(subset_data@meta.data,
aes(x=group, fill=Idents(subset_data))) + geom_bar(position = "fill")
#######3 cellchat数据准备 ---------
subset_data[["percent.mt"]] <- PercentageFeatureSet(subset_data, pattern = "^mt-")
VlnPlot(subset_data, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
library(dplyr)
subset_data = subset_data %>%
Seurat::NormalizeData(verbose = FALSE) %>%
FindVariableFeatures(selection.method = "vst", nfeatures = 2000) %>%
ScaleData(verbose = FALSE) %>%
RunPCA(npcs = 50, verbose = FALSE)
ElbowPlot(subset_data, ndims = 50)
VlnPlot(subset_data, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
## 466 510
#subset_data@meta.data$stim <- c(rep("Exp", length(grep("1$",colnames(subset_data)))),rep("Con", length(grep("2$",colnames(subset_data)))))
#table(subset_data$stim)
library('harmony')
subset_data <- subset_data %>% RunHarmony("stim", plot_convergence = TRUE)
harmony_embeddings <- Embeddings(subset_data, 'harmony')
#######################cluster
dims = 1:15
subset_data <- subset_data %>%
RunUMAP(reduction = "harmony", dims = dims) %>%
RunTSNE(reduction = "harmony", dims = dims) %>%
FindNeighbors(reduction = "harmony", dims = dims)
subset_data=FindClusters(subset_data )
DimPlot(subset_data,label = TRUE,repel = TRUE)|DimPlot(subset_data,label = TRUE,group.by = "cell.type")
DimPlot(subset_data,label = TRUE,repel = TRUE,reduction = 'tsne')
head(subset_data@meta.data)
#########cellchat-------
#library(dplyr)
library(cowplot)
library(Seurat)
library(harmony)
library(ggplot2)
require(Matrix)
require(magrittr)
library(openxlsx)
#BiocManager::install('CellChat',force = TRUE)
#devtools::install_github("jinworks/CellChat",dependencies = TRUE)
library(CellChat)
getwd()
table(Idents(subset_data))
table(subset_data$group)
#subset_data$stim=subset_data$group
print(getwd())
DimPlot(subset_data,label =TRUE,group.by = 'cell.type',reduction = 'tsne')
table(Idents(subset_data))
Idents(subset_data)=subset_data$cell_type
cellchat分析(批量运行代码)
path=getwd()
#head(subset_data@meta.data)
#Idents(subset_data)
#table(subset_data$stim)
## CellChat
dir.create(paste(path, "CellChat_stim", sep = "/"))
setwd(paste(path, "CellChat_stim", sep = "/"))
getwd()
#setwd("../")
table(subset_data$group)
table(subset_data$stim)
subset_data$stim=subset_data$group
# setwd("../")
getwd()
library(parallel)
library(CellChat)
library(openxlsx)
# 获取 unique(stim) 的值
stim_list <- unique(subset_data$stim)
# 定义并行处理函数
process_stim <- function(stim) {
# stim='Bleomycin'
print(stim);getwd()
dir.create(paste(path, "cellchat", stim, sep = "/"), recursive = TRUE)
setwd(paste(path, "cellchat", stim, sep = "/"))
data.input <- GetAssayData(subset_data, slot = "data")[, subset_data$stim == stim]
meta <- data.frame(labels = Idents(subset_data)[subset_data$stim == stim], row.names = colnames(subset_data)[subset_data$stim == stim])
meta$labels <- droplevels(meta$labels, exclude = setdiff(levels(meta$labels), unique(meta$labels)))
cellchat <- createCellChat(object = data.input, meta = meta, group.by = "labels")
cellchat@DB <- CellChatDB.mouse
cellchat <- subsetData(cellchat)
cellchat <- identifyOverExpressedGenes(cellchat)
cellchat <- identifyOverExpressedInteractions(cellchat)
cellchat <- projectData(cellchat, PPI.mouse)
cellchat <- computeCommunProb(cellchat)
cellchat <- filterCommunication(cellchat, min.cells = 0)
cellchat <- computeCommunProbPathway(cellchat)
cellchat <- aggregateNet(cellchat)
df.net <- subsetCommunication(cellchat)
write.xlsx(df.net, file = "0.Cell-Cell_Communications_At_L-R.xlsx", rowNames = FALSE, colNames = TRUE)
df.net <- subsetCommunication(cellchat, slot.name = "netP")
write.xlsx(df.net, file = "0.Cell-Cell_Communications_At_Pathway.xlsx", rowNames = FALSE, colNames = TRUE)
groupSize <- as.numeric(table(cellchat@idents))
mat <- cellchat@net$count
write.xlsx(mat, file = "1.NumberOfInteractions.xlsx", rowNames = TRUE, colNames = TRUE)
pdf("1.NumberOfInteractions.pdf")
netVisual_circle(mat, vertex.weight = groupSize, weight.scale = TRUE, label.edge = FALSE, title.name = "Number of interactions")
dev.off()
mat <- cellchat@net$weight
write.xlsx(mat, file = "2.InteractionWeights.xlsx", rowNames = TRUE, colNames = TRUE)
pdf("2.InteractionWeights.pdf")
netVisual_circle(mat, vertex.weight = groupSize, weight.scale = TRUE, label.edge = FALSE, title.name = "Interaction weights/strength")
dev.off()
save(cellchat, file = paste0("cellchat_", stim, "_.RData"))
pdf(paste0("3.Sig_Pathway_Hierarchy_Plot_",stim,"_.pdf"))
for(i in pathways){
#i=1
print(i)
p = netVisual_aggregate(cellchat, signaling = i,vertex.receiver = vertex.receiver,
vertex.label.cex = 0.4,layout = 'hierarchy')
title(main = paste0(i,' signaling'))
print(p)
}
dev.off()
getwd()
# a=1
# a
# getwd()
# setwd("./cellchat/SiO2_7/")
# load("./cellchat/SiO2_7/cellchat_SiO2_7_.RData")
# pathways = cellchat@netP$pathways
#
library(CellChat)
pdf(paste0("3.Sig_Pathway_Circle_Plot_",stim,"_.pdf"))
for(i in pathways){
print(i)
p = netVisual_aggregate(cellchat,show.legend = TRUE,
signaling = i,signaling.name = paste(i,"sigaling"),
,pt.title=10,vertex.receiver = vertex.receiver, layout = "circle")
title(main = paste0(i,' signaling'))
print(p)
}
dev.off()
pdf(paste0("4.Sig_Pathway_L-R_pair_Contribution_",stim,"_.pdf"))
for(i in pathways){
print(i)
p = netAnalysis_contribution(cellchat, signaling = i, title = paste0(i, " signaling pathway", " Contribution of each L-R pair"))
print(p)
}
dev.off()
pdf(paste0("4.Sig_Pathway_L-R_pair_bubbleplot_",stim,"_.pdf"), width=25, height=20)
p = netVisual_bubble(cellchat, remove.isolate = FALSE)
print(p)
dev.off()
cellchat <- netAnalysis_computeCentrality(cellchat, slot.name = "netP")
pdf(paste0("5.Signaling_Roles_Of_Cell_Groups_Heatmap_",stim,"_.pdf"))
for(i in pathways){
print(i)
p = netAnalysis_signalingRole_network(cellchat, signaling = i, width = 8, height = 2.5, font.size = 10)
print(p)
}
dev.off()
pdf(paste0("4.Sig_Pathway_L-R_pair_bubbleplot_",stim,"_.pdf"), width=25, height=20)
p = netVisual_bubble(cellchat, remove.isolate = FALSE)
print(p)
dev.off()
pdf(paste0("5.Signaling_Roles_Of_Cell_Groups_2D_",stim,"_.pdf"))
p = netAnalysis_signalingRole_scatter(cellchat)
print(p)
dev.off()
pdf(paste0("5.signals_Contribution_Of_Cell_Groups_Heatmap_",stim,"_.pdf"), width=10)
ht1 <- netAnalysis_signalingRole_heatmap(cellchat, pattern = "outgoing", font.size = 5)
ht2 <- netAnalysis_signalingRole_heatmap(cellchat, pattern = "incoming", font.size = 5)
print(ht1 + ht2)
dev.off()
save(cellchat, file = paste0("cellchat_",stim,"_.RData"))
return(NULL)
}
# 设置并行计算
num_cores <- 2 #detectCores() - 1 # 使用所有可用核心的 n-1
mclapply(stim_list, process_stim, mc.cores = num_cores)


cellchat两组比较
#request 2
.libPaths(c( "/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2",
"/home/data/t040413/R/yll/usr/local/lib/R/site-library",
"/refdir/Rlib/", "/usr/local/lib/R/library"))
getwd()
filepath="~/biye/ipf/gse104154/cellchat/merge"
dir.create(filepath,recursive = TRUE)
setwd(filepath)
list.dirs()
load("~/biye/ipf/gse104154/cellchat/cellchat/Control/cellchat_Control_.RData")
ns=cellchat
load("~/biye/ipf/gse104154/cellchat/cellchat/Bleomycin/cellchat_Bleomycin_.RData")
sio2=cellchat
#两个数据集的细胞类型一致情况下的:联合分析---
#https://htmlpreview.github.io/?https://github.com/jinworks/CellChat/blob/master/tutorial/Comparison_analysis_of_multiple_datasets.html
object.list <- list(Pbs = ns, Bleomycin = sio2)
cellchat <- mergeCellChat(object.list, add.names = names(object.list))
#> Merge the following slots: 'data.signaling','images','net', 'netP','meta', 'idents', 'var.features' , 'DB', and 'LR'.
cellchat
ptm = Sys.time();ptm
execution.time = Sys.time() - ptm
print(as.numeric(execution.time, units = "secs"))
getwd()
#1保存merge数据----
# Users can now export the merged CellChat object and the list of the two separate objects for later use
save(object.list, file = "cellchat_object.list_humanSkin_NL_LS.RData")
save(cellchat, file = "cellchat_merged_humanSkin_NL_LS.RData")
#1.1整体比较---
ptm = Sys.time()
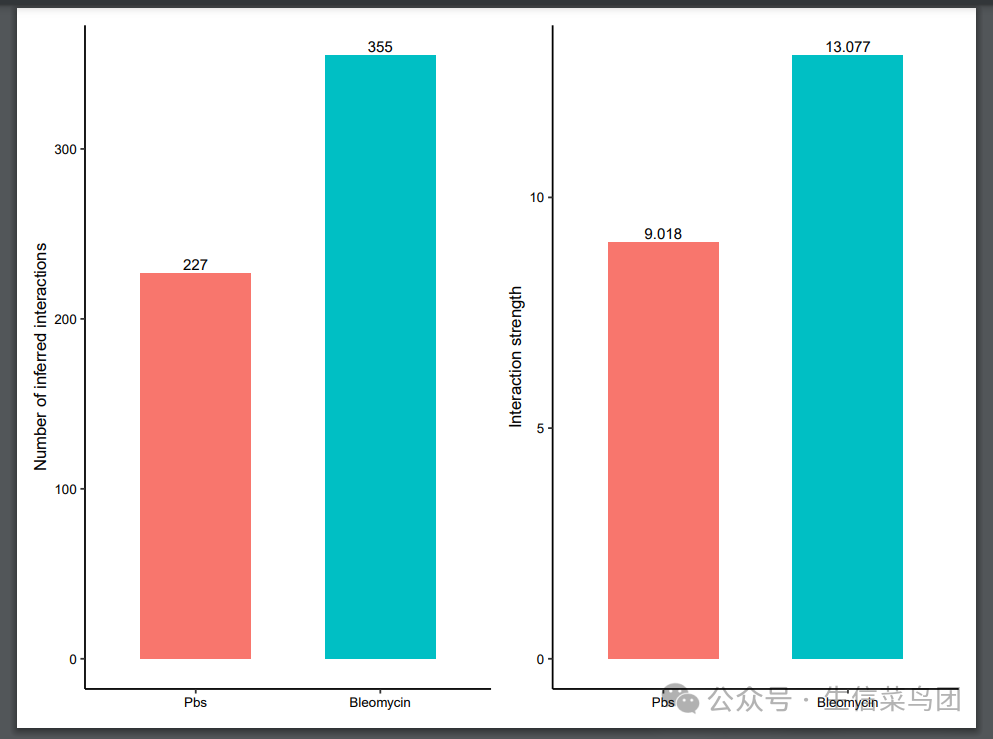
gg1 <- compareInteractions(cellchat, show.legend = F, group = c(1,2))
gg2 <- compareInteractions(cellchat, show.legend = F, group = c(1,2), measure = "weight")
gg1 + gg2
ggsave(plot = gg1 + gg2,filename = "1.1整体比较.pdf",height =6,width = 8,limitsize = FALSE)
getwd()
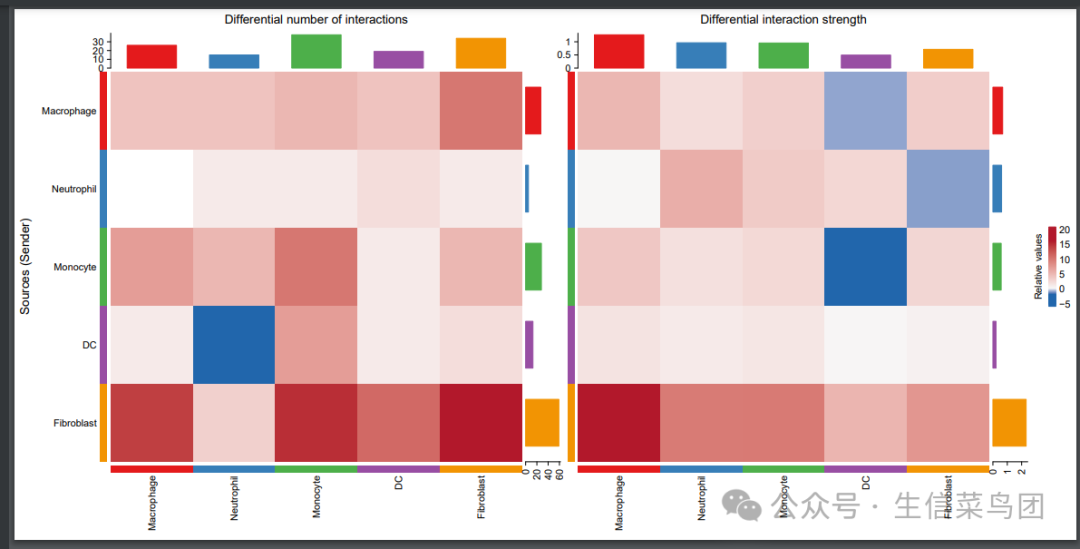
#1.2个数和强度比较弦图---
# red(or blue) colored edges represent increased(or decreased) signaling in the second dataset compared to the first one.
pdf("1.2个数和强度比较弦图.pdf",height = 6,width = 12)
par(mfrow = c(1,2), xpd=TRUE)
layout(matrix(c(1,2), nrow = 1, ncol = 2, byrow = TRUE))
print(netVisual_diffInteraction(cellchat, weight.scale = T))
print(netVisual_diffInteraction(cellchat, weight.scale = T, measure = "weight"))
dev.off( )
#1.2.2 个数和强度比较热图---
gg1 <- netVisual_heatmap(cellchat)
#> Do heatmap based on a merged object
gg2 <- netVisual_heatmap(cellchat, measure = "weight")
#> Do heatmap based on a merged object
gg1 + gg2
pdf("1.2.2 个数和强度比较热图.pdf",height = 6,width = 12)
layout(matrix(c(1,2), nrow = 1, ncol = 2, byrow = TRUE))
print(gg1+gg2)
dev.off( )
num.link <- sapply(object.list, function(x) {rowSums(x@net$count) + colSums(x@net$count)-diag(x@net$count)})
weight.MinMax <- c(min(num.link), max(num.link)) # control the dot size in the different datasets
gg <- list()
for (i in 1:length(object.list)) {
gg[[i]] <- netAnalysis_signalingRole_scatter(object.list[[i]], title = names(object.list)[i], weight.MinMax = weight.MinMax)
}
#> Signaling role analysis on the aggregated cell-cell communication network from all signaling pathways
#> Signaling role analysis on the aggregated cell-cell communication network from all signaling pathways
patchwork::wrap_plots(plots = gg)
ggsave(plot = patchwork::wrap_plots(plots = gg),filename = "1.2.3_outgoing_incoming.pdf",width = 12,height = 6,limitsize = FALSE)
gg1 <- netAnalysis_signalingChanges_scatter(cellchat, idents.use = "Fibroblast", signaling.exclude = "MIF")
#> Visualizing differential outgoing and incoming signaling changes from NL to LS
#> The following `from` values were not present in `x`: 0
#> The following `from` values were not present in `x`: 0, -1
gg2 <- netAnalysis_signalingChanges_scatter(cellchat, idents.use = "Fibroblast", signaling.exclude = c("MIF"))
#> Visualizing differential outgoing and incoming signaling changes from NL to LS
#> The following `from` values were not present in `x`: 0, 2
#> The following `from` values were not present in `x`: 0, -1
patchwork::wrap_plots(plots = list(gg1,gg2))
#2.1功能相似--------
ptm = Sys.time()
cellchat <- computeNetSimilarityPairwise(cellchat, type = "functional")
#> Compute signaling network similarity for datasets 1 2
cellchat <- netEmbedding(cellchat, type = "functional")
#> Manifold learning of the signaling networks for datasets 1 2
cellchat <- netClustering(cellchat, type = "functional")
#> Classification learning of the signaling networks for datasets 1 2
# Visualization in 2D-space
netVisual_embeddingPairwise(cellchat, type = "functional", label.size = 3.5)
#> 2D visualization of signaling networks from datasets 1 2
#netVisual_embeddingZoomIn(cellchat, type = "functional", nCol = 2)
#2.2结构相似----
cellchat <- computeNetSimilarityPairwise(cellchat, type = "structural")
cellchat <- netEmbedding(cellchat, type = "structural")
cellchat <- netClustering(cellchat, type = "structural")
# Visualization in 2D-space
netVisual_embeddingPairwise(cellchat, type = "structural", label.size = 3.5)
#netVisual_embeddingPairwiseZoomIn(cellchat, type = "structural", nCol = 2)
#2.3两个数据集之间的通路差异距离----
rankSimilarity(cellchat, type = "functional")
#> Compute the distance of signaling networks between datasets 1 2
#比较信息流-----
gg1 <- rankNet(cellchat, mode = "comparison", measure = "weight", sources.use = NULL, targets.use = NULL, stacked = T, do.stat = TRUE)
gg2 <- rankNet(cellchat, mode = "comparison", measure = "weight", sources.use = NULL, targets.use = NULL, stacked = F, do.stat = TRUE)
gg1 + gg2
ggsave(plot = patchwork::wrap_plots(plots = gg1 + gg2),
filename = "14_information_flow.pdf",width = 12,height = 6,limitsize = FALSE)
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-02-08,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读

