【再复杂的实验设计也不怕】单细胞数据pseudobulk及差异分析集成包dreamlet
【再复杂的实验设计也不怕】单细胞数据pseudobulk及差异分析集成包dreamlet

KS科研分享与服务-TS的美梦
发布于 2025-12-20 16:57:26
发布于 2025-12-20 16:57:26
之前介绍完单细胞数据pseudobulk分析之后(参考:玩转单细胞(23):基于seurat包AggregateExpression函数的单细胞转录组pseudobulk分析、muscat: 单细胞转录组pseudobulk分析R包更新),有小伙伴问过自己的实验设计非常复杂,不是简单的两组比较,如何操作,只能按照正常的进行两两分组比较了。之后在完整复现Nature子刊(Nature immunology)文章主图Figure2全部内容这篇Nature子刊的学习中接触到一个新的包---dreamlet,能够应对更加复杂的实验设计,完成单细胞数据pseudobulk分析以及差异分析!
dreamlet简介及安装
dreamlet(也算是对muscat的拓展)是一个用于单细胞转录组学数据的可扩展差异表达分析工具包,也就是将pseudobulk分析以及后续差异分析整个流程完成;它专为具有复杂实验设计的多样本单细胞数据集而开发(比如目前不仅是测序量大,包括数据库整合,样本多,实验设计也更加复杂,不仅只有正常组实验组之间的简单设计,更复杂的包含了性别、年龄、药物、基因编辑等等条件,这对于计算有很大的要求,Dreamlet则进一步扩展了对大规模单细胞/单核转录组数据集的分析能力,通过优化计算效率与内存管理,有效解决了传统分析流程中CPU运算与内存使用的瓶颈问题)。当然,一般数据集分析也是可以使用的,只不过无需使用那么多复杂的功能而已!
Github:https://github.com/GabrielHoffman/dreamlet?tab=readme-ov-file Paper:https://www.biorxiv.org/content/10.1101/2023.03.17.533005v2
#dreamlet >= 1.0.0 is compatible with BioC v3.18 for R v4.3.
#devtools::install_github("DiseaseNeurogenomics/dreamlet")
#or
#BiocManager::install("zellkonverter")
library(dreamlet)
package.version("dreamlet")
## [1] "1.8.0"示例数据
演示数据来源于https://www.nature.com/articles/s41590-025-02241-4#data-availability这篇NI文章(完整复现Nature子刊(Nature immunology)文章主图Figure2全部内容)。读入数据:dreamlet的input data需要是SingleCellExperiment对象,如果您的数据是seurat,那么很好办,as.SingleCellExperiment()即可完成转化;如果您的数据是python h5ad对象,可以像这里演示的一样,直接使用zellkonverter包的readH5AD函数读取,并会直接转化为SingleCellExperiment object。当然,还有方法是将h5ad转化为seurat或者SingleCellExperiment,参考:https://mp.weixin.qq.com/s/x4HOHNwikvuTCRK4kQRjRA;总之,办法很多。
setwd('~/data_analysis/dreamlet伪bulk分析/')
library(SingleCellExperiment)
library(zellkonverter)sce <- readH5AD('./lymphoid.h5ad', reader = "R", #R or python,python使用reticulate读取,可能会出现不必要的麻烦error,使用R用zellkonverter's native R reader.
use_hdf5=TRUE, verbose = TRUE)
sce
## class: SingleCellExperiment
## dim: 35475 130414
## metadata(9): IA_norm_citeseq IA_raw_citeseq ... schema_version title
## assays(1): X
## rownames(35475): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
## ENSG00000277196
## rowData names(6): feature_is_filtered feature_name ... feature_length
## feature_type
## colnames(130414): AAACCCAAGCATGTTC-1_CZINY-0061-12
## AAACCCAAGCCAGAGT-1_CZINY-0052-3 ... TTTGTTGTCGGAACTT-1_CZINY-0063-13
## TTTGTTGTCGTTAGAC-1_CZINY-0060-11
## colData names(46): reference_genome gene_annotation_version ...
## development_stage observation_joinid
## reducedDimNames(1): X_umap
## mainExpName: NULL
## altExpNames(0): 看看示例数据metadata:示例数据有130414细胞,是亚群数据集合,所以不是特别大。这个实验设计是比较复杂的,首先是来源于不同的组织,且sample还有性别、年龄.
#celltype annotation
sce$author_cell_type <- as.character(sce$author_cell_type)
sce$celltype <- sce$author_cell_type
sce$celltype[sce$author_cell_type %in% c("nk_cd56dim")] <- "CD56dim NK"
sce$celltype[sce$author_cell_type %in% c("ilc_3")] <- "ILC3"
sce$celltype[sce$author_cell_type %in% c("nk_ilc_precursor")] <- "Pre NK/ILC"
sce$celltype[sce$author_cell_type %in% c("ilc_1","ilc_1_proliferating","ilc_1/3_transitional")] <- "ILC1"
sce$celltype[sce$author_cell_type %in% c("nk_cd56br","nk_cd56dim_proliferating","nk_cd56br_proliferating")] <- "CD56br NK"
# 将 celltype 设为当前分组标签
colLabels(sce) <- sce$celltype
plotProjection(sce, "X_umap", "celltype", legend.position="none", text=TRUE)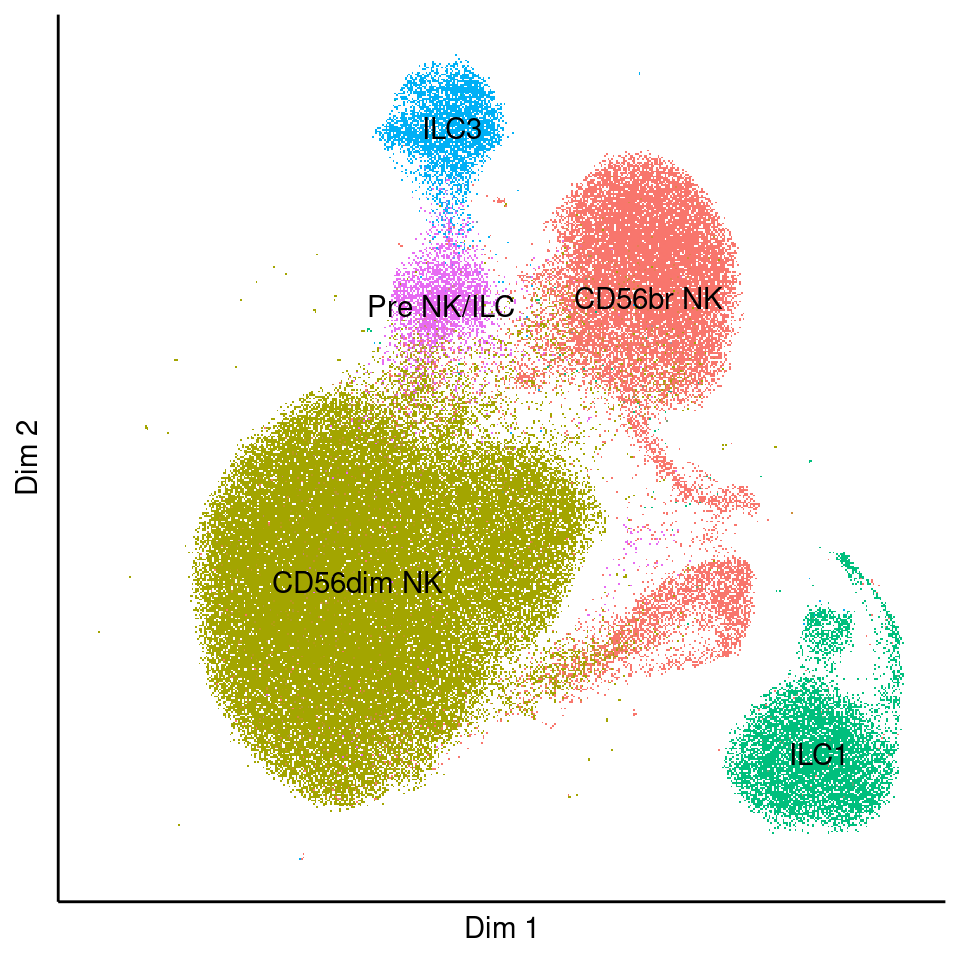
#年龄分为两组,>40 & <40
sce$Age <- sce$donor_age
sce$Age[sce$donor_age %in% c("67 years","56 years","75 years","68 years","50-54 years","64 years","48 years","40-44 years","70-74 years", "55-60 years","73 years")] <- ">40"
sce$Age[sce$donor_age %in% c("23 years","33 years","20-24 years","25 years","27 years","20 years","35-39 years","25-30 years")] <- "<40"#修改下组织名称
sce$tissue <- recode(
sce$tissue,
"bone marrow" = "BM",
"blood" = "BL",
"spleen" = "SP",
"jejunal epithelium" = "JE",
"epithelial lining fluid" = "ELF",
"lung" = "LG",
"jejunum lamina propria" = "JLP",
"thoracic lymph node" = "TLN",
"mesenteric lymph node" = "MLN",
"inguinal lymph node" = "ILN"
)
plotProjection(sce, "X_umap", "tissue", legend.position="right", text=FALSE)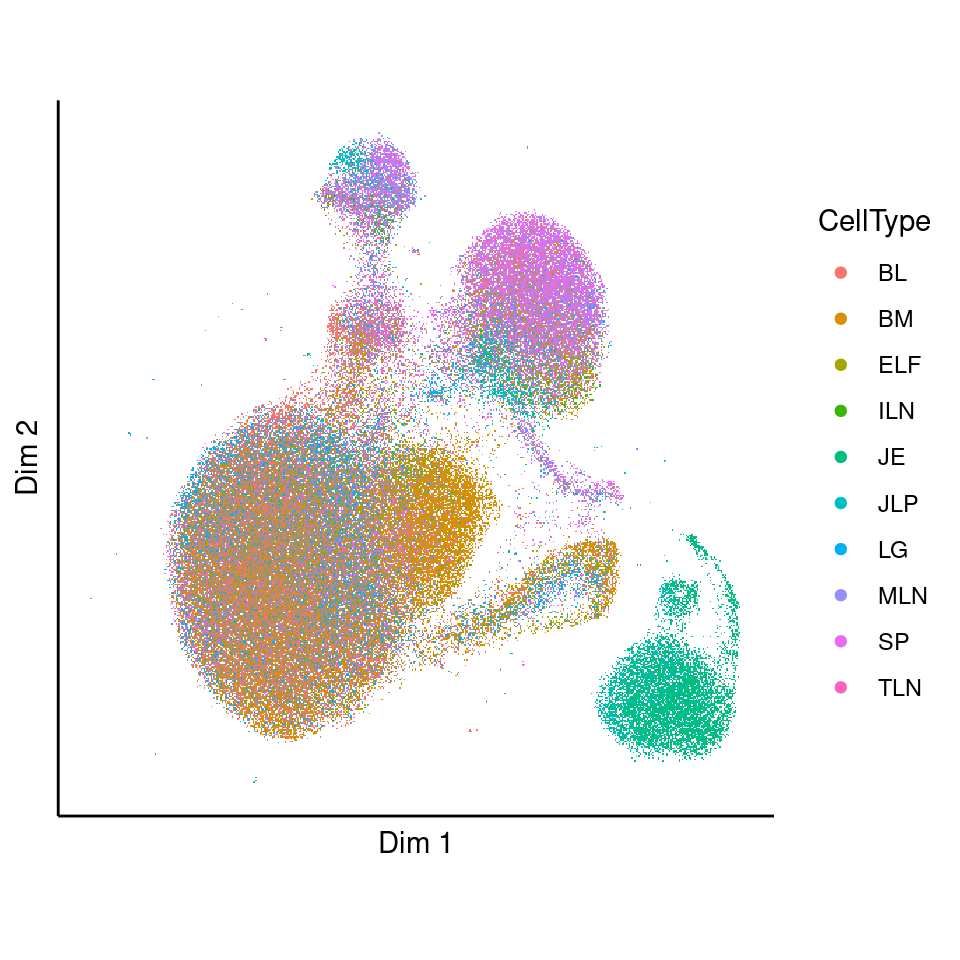
Aggregate to pseudobulk
Dreamlet与muscat包类似,通过将给定样本和细胞类型的原始计数在细胞层面求和来进行伪bulk水平的分析。对于大型磁盘数据集,aggregateToPseudoBulk比muscat::aggregateData快得多。dreamlet函数aggregateToPseudoBulk通过指定cluster_id和sample_id创建伪批量数据,然后将每种细胞类型的计数数据存储在“assay filed”。
#合并tissue与id,是的每个id在每个组织中有唯一标识
sce$donor_tissue <- paste0(sce$tissue, "_",sce$donor_id)
pb <- aggregateToPseudoBulk(
sce,
assay = "X",
cluster_id = "celltype",
sample_id = "donor_tissue",
verbose = TRUE)Voom for pseudobulk
使用voomWithDreamWeights对每种celltype内的伪bulk counts应用voom式标准化,以处理随机效应,然后进行后续的差异分析。processAssays()函数会保留给定细胞类型中至少包含 min.cells个细胞的样本。虽然通常丢弃少量样本不是问题,但在某些情况下,丢弃样本可能意味着回归公式中包含的某个变量不再具有任何变化。实际情况中,如果某个样本包含很少的细胞数,那么不太适合纳入分析!
#多线程运算
library(BiocParallel)
multicoreParam <- MulticoreParam(workers = 5)
multicoreParam# Normalize and apply voom/voomWithDreamWeights
res.proc = processAssays(pb, ~ tissue + donor_age + sex + cmv, min.cells=10, min.count=5, min.prop=0.1, BPPARAM=multicoreParam)#展示了通过voom方法得到的各细胞类型的均值–方差(mean–variance)关系趋势。
# show voom plot for each cell clusters
options(repr.plot.width = 100, repr.plot.height = 16, repr.plot.res = 100)
plotVoom(res.proc)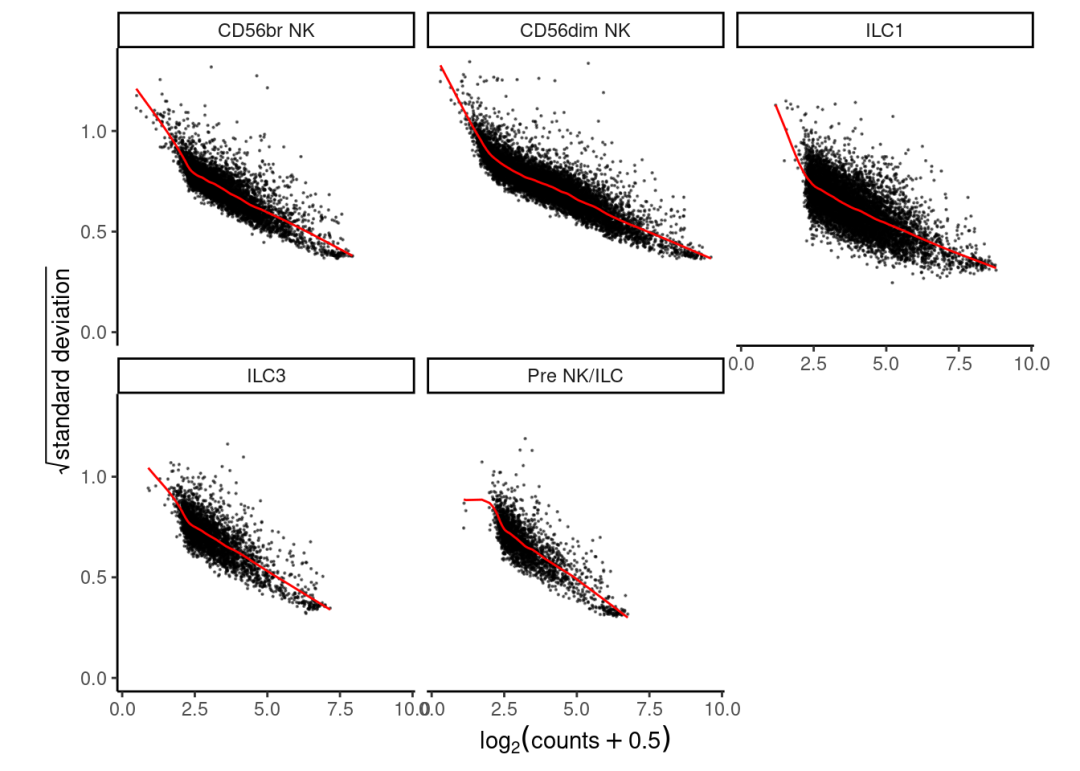
差异分析
标准化的表达数据和metadata都存储在processAssays预处理后的res.proc中,之后需要指定回归公式比较组进行差异分析,差异分析使用的函数是dreamlet,会对每一个assay(in res.proc)使用线性/线性混合模型执行差异表达分析。如果您的实验设计简单,就是常见的实验组、对照组,那么仅仅需要指定实验设计状态即可,使用线性模型,例如官网教程中(res.dl <- dreamlet(res.proc, ~StimStatus))。但是,对于更大规模数据集的分析可以包含协变量和随机效应,比如这个示例中,年龄、性别都是固定效应,原文中还有其他的随机效应(比如form <- ~ tissue_groups + cmv + age_group + sex + (1|chemistry) + (1|site) + 0),定义了一个线性混合模型(Linear Mixed Model)公式,用于后续拟合每个基因的表达。
colData(res.proc)$tissue_sex <- as.factor(paste0(colData(res.proc)$tissue,'_',colData(res.proc)$sex))
form <- ~ tissue_sex + cmv + Age + 0
# Define contrasts
# Note that for each contrass, the weights sum to 1
L <- makeContrastsDream(form, colData(res.proc), contrasts = c(
BM_sex = "tissue_sexBM_male - tissue_sexBM_female",
BL_sex = "tissue_sexBL_male - tissue_sexBL_female",
SP_sex = "tissue_sexSP_male - tissue_sexSP_female",
JE_sex = "tissue_sexJE_male - tissue_sexJE_female")
)
# Plot to visualize contrasts matrix
plotContrasts(L)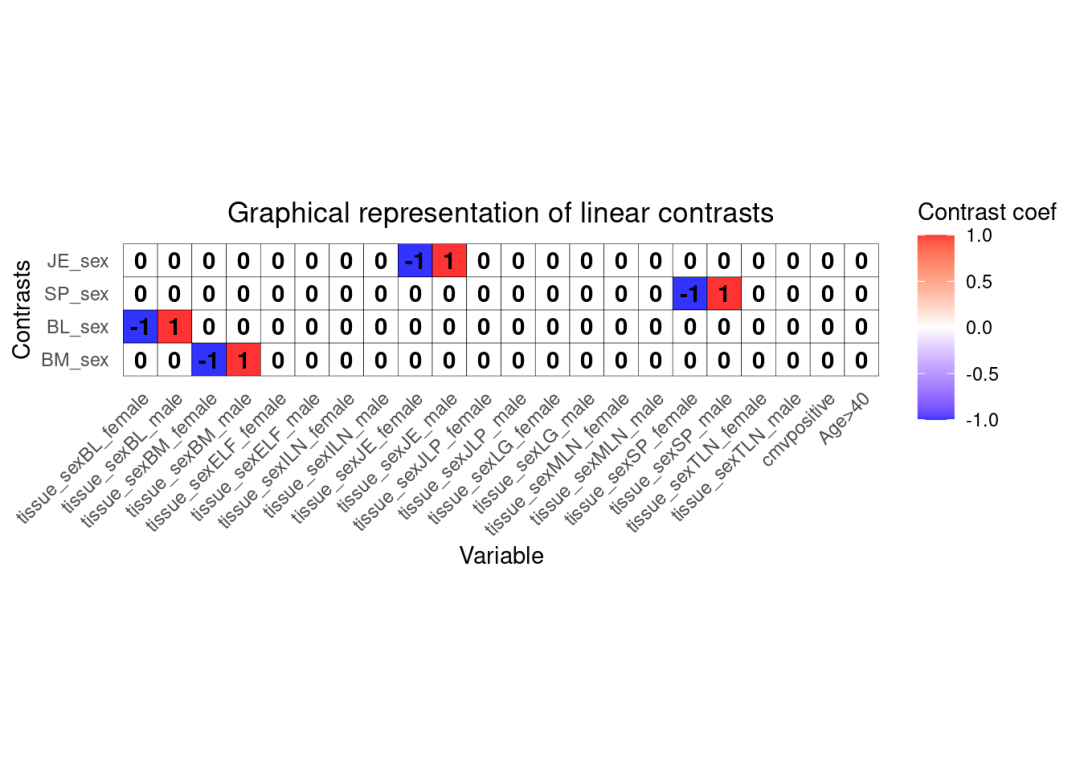
差异分析:
# Differential expression analysis within each assay,
# evaluated on the voom normalized data
res.dl2 <- dreamlet(
x=res.proc,#SingleCellExperiment or dreamletProcessedData object
formula=form,
assays= assayNames(res.proc),
data = colData(res.proc),
contrasts = c(
BM_sex = "tissue_sexBM_male - tissue_sexBM_female",
BL_sex = "tissue_sexBL_male - tissue_sexBL_female",
SP_sex = "tissue_sexSP_male - tissue_sexSP_female",
JE_sex = "tissue_sexJE_male - tissue_sexJE_female"),#比较组
#computeResiduals=TRUE,
#BPPARAM=multicoreParam
)火山图查看差异分析结果
上述的差异分析比较,执行的是不同组织中,不同celltype中性别差异基因:
plotVolcano(res.dl2, coef = "BM_sex",nGenes = 10,ncol = 5)#nGenes需要在途中highlight显著表达基因数目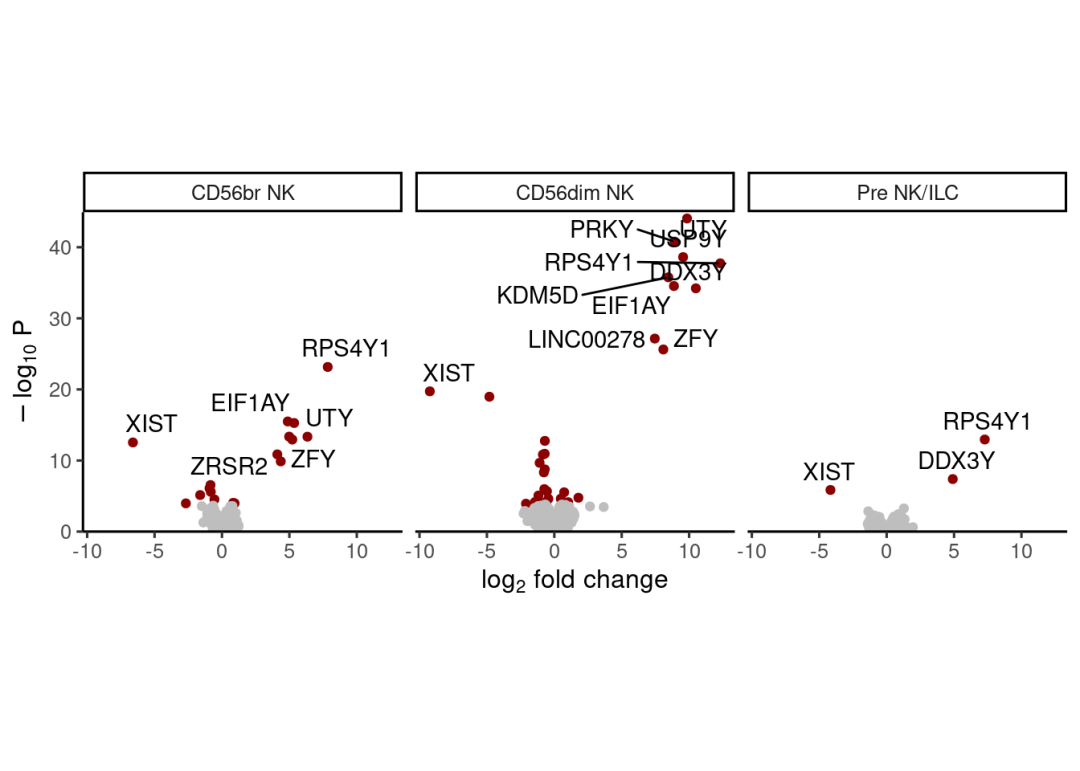
plotVolcano(res.dl2, coef = "BL_sex",nGenes = 10,ncol = 5)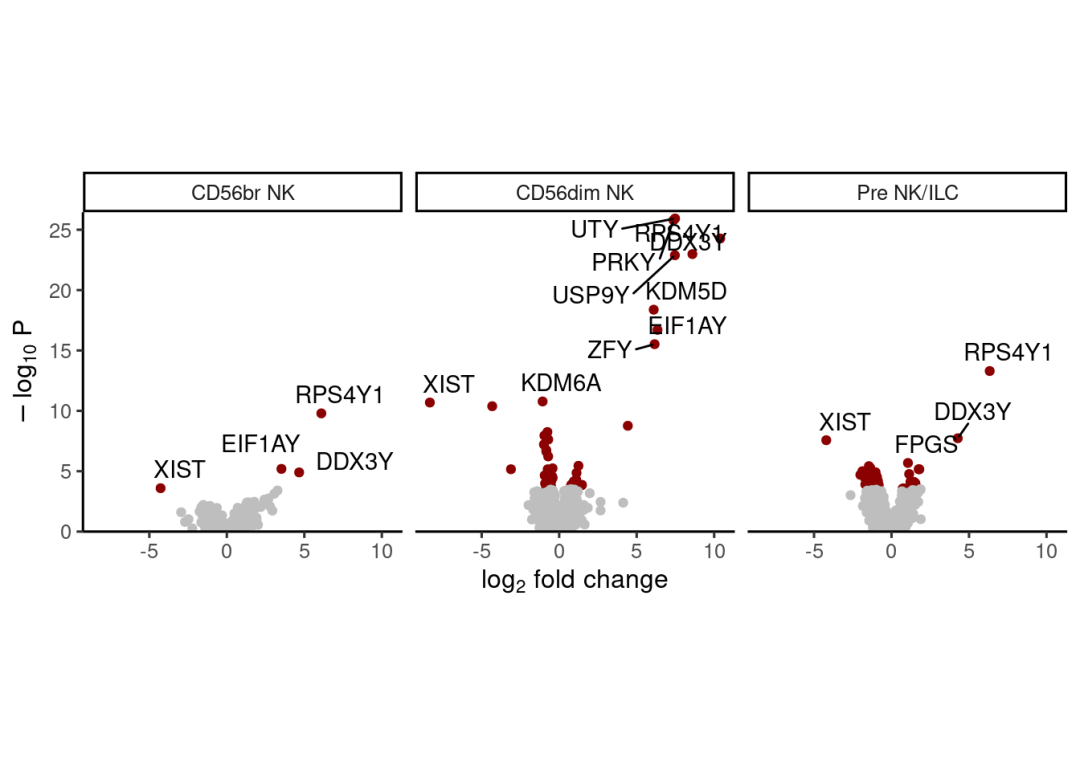
plotVolcano(res.dl2, coef = "SP_sex",nGenes = 10,ncol = 5)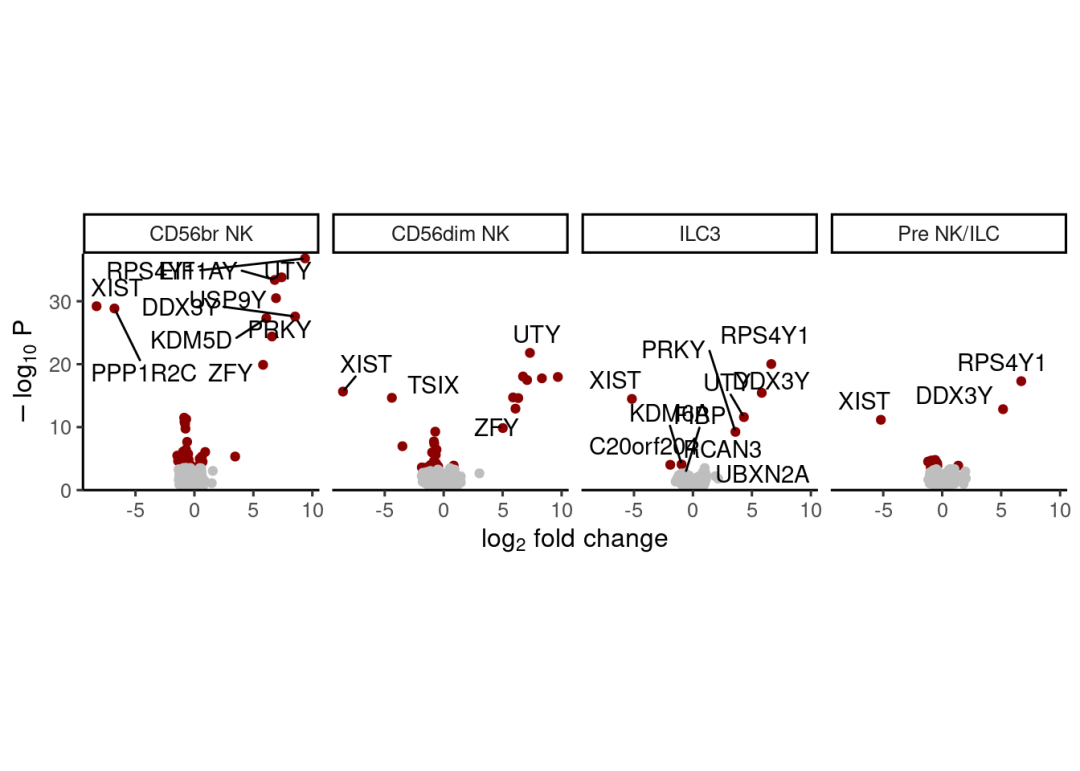
plotVolcano(res.dl2, coef = "JE_sex",nGenes = 10,ncol = 5)
提取分析结果
保存所有结果:
# for (i in c("BM_sex","BL_sex","SP_sex","JE_sex")){
# table <- topTable(res.dl2, coef=i, number=Inf)
# write.csv(table, paste0( i, "_male_vs_female.csv"))
# }以上就是全部内容了,官网教程中还有关于dreamlet包差异分析后续的富集分析以及可视化,我想这不是这个包的重点,也并非是必须的,得到差异结果才是我们的目的,这个结果可以进行其他各种可视化以及富集分析!
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-11-20,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

