画个同款新奇的“Galaxy”星系UMAP图(Nat Immunol:IF27.8)
画个同款新奇的“Galaxy”星系UMAP图(Nat Immunol:IF27.8)
生信技能树
发布于 2025-02-25 16:09:40
发布于 2025-02-25 16:09:40
前面我们已经介绍了如何在umap图上加圈:给你的单细胞umap图加个cell杂志同款的圈,以及绘制星系umap图:5种方式美化你的单细胞umap散点图,那两者组合起来呢?今天学习的这个好看的图来自文献《The aged tumor microenvironment limits T cell control of cancer》,于2024年6月25日发表在Nat Immunol杂志上(IF27.8)。如下:
颜色的深浅表示细胞密度,圈内为不同的细胞类型,左边为年轻组,右边为老年组,可以看出 yong 和 old组两种截然不同的细胞浸润模式。
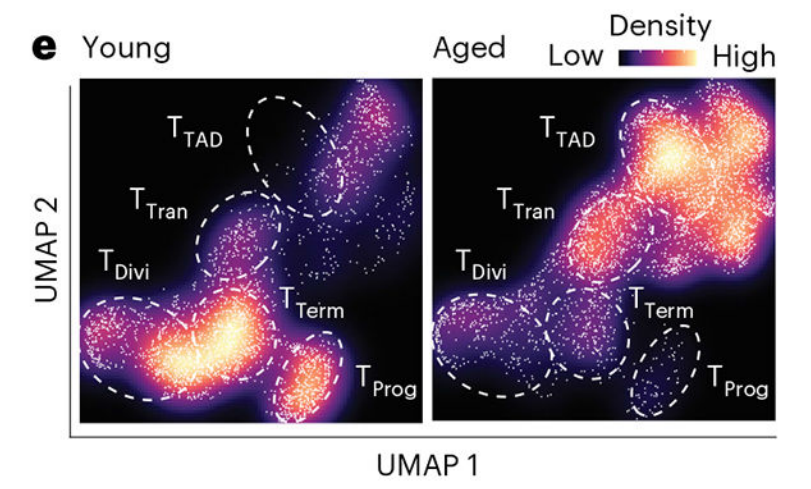
图注:
Fig. 1 ∣. Aging promotes tumor growth and alters CD8+ T cell fate and effector function. . e, Galaxy plots depicting the cell density in UMAP space for tumor-infiltrating CD8+ T cells from young (left) and aged (right) tumor-bearing mice. Cooler colors indicate low density, and warmer colors indicate high density. The relative proportion of CD8+ T cells from young (blue, n = 5) and aged (red, n = 5) tumors within TProg, TTerm, dividing T (TDivi) and TTAD cell clusters is shown。
示例数据
使用的数据还是自 GSE128531 数据注释后的seurat对象,你自己用的时候可以使用任何一个经过了注释后的seurat对象。
###
### Create: Jianming Zeng
### Date: 2023-12-31
### Email: jmzeng1314@163.com
### Blog: http://www.bio-info-trainee.com/
### Forum: http://www.biotrainee.com/thread-1376-1-1.html
### CAFS/SUSTC/Eli Lilly/University of Macau
### Update Log: 2023-12-31 First version
### Update Log: 2024-12-09 by juan zhang (492482942@qq.com)
###
rm(list=ls())
library(COSG)
library(harmony)
library(ggsci)
library(dplyr)
library(future)
library(Seurat)
library(clustree)
library(cowplot)
library(data.table)
library(dplyr)
library(ggplot2)
library(patchwork)
library(stringr)
library(qs)
# 导入数据
sce.all.int <- readRDS('2-harmony/sce.all_int.rds')
sp <- 'human'
head(sce.all.int@meta.data)
load("phe.Rdata")
head(phe)
sce.all.int <- AddMetaData(sce.all.int, metadata = phe)
Idents(sce.all.int) <- "celltype"
先画银河中星系
用的 ggpointdensity 包,绘制不同的细胞密度:5种方式美化你的单细胞umap散点图
library(ggpointdensity)
# 提取数据
# dat <- Embeddings(sce.all.int, reduction = "umap")
# head(dat)
dat <- FetchData(object=sce.all.int, vars=c("umap_1","umap_2","celltype"))
head(dat)
p <- ggplot(data = dat, mapping = aes(x = umap_1, y = umap_2)) +
geom_pointdensity() +
scale_color_viridis_c(option="inferno", alpha = 0.4) +
theme_classic(base_size = 15)
p
结果如下:

添加银河黑暗背景
这里简单的调整ggplot主题:
p1 <- p +
labs(color='Density') + # 设置图例标题
theme(
panel.background = element_rect(fill = "black", color = "black"), # 设置坐标轴内区域背景颜色为黑色
plot.background = element_rect(fill = "white", color = "white"), # 设置整个图形背景颜色为白色
panel.grid.major = element_blank(), # 去掉主要的网格线
panel.grid.minor = element_blank(), # 去掉次要的网格线
axis.line = element_line(color = "white"), # 设置坐标轴线颜色为白色
axis.text = element_text(color = "white"), # 设置坐标轴文本颜色为白色
axis.ticks = element_line(color = "white"), # 设置坐标轴刻度颜色为白色
axis.title = element_text(color = "white"), # 设置坐标轴标题文本颜色为白色
legend.background = element_blank(), # 设置图例背景为透明
legend.key = element_blank(), # 设置图例键为透明
legend.text = element_text(color = "white"), # 设置图例文本颜色为白色
legend.title = element_text(color = "black")) # 设置图例标题文本颜色为白色
p1
结果如下:
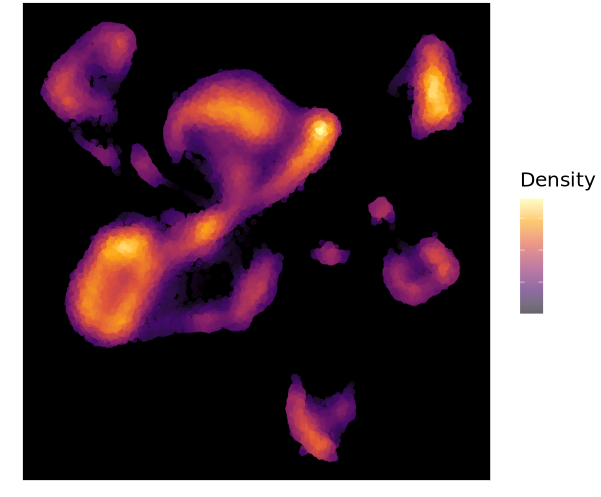
加圈圈住
上次介绍的办法:mascarade 包,给你的单细胞umap图加个cell杂志同款的圈
# 加圈
library(mascarade)
# 制作masktable
# smoothSigma = 0.05:控制加圈的平滑成都,值越大加的圈越平滑
# minDensity :控制 加圈的松紧成都,值越小,加的圈边界与umap散点距离越大越宽松
maskTable <- generateMask( dims=dat[,1:2], cluster=dat$celltype, minDensity = 15,smoothSigma = 0.1 )
class(maskTable)
dim(maskTable)
head(maskTable)
p2 <- p1 +
geom_path(data=maskTable, aes(group=group),linewidth=0.6,linetype = 2, colour = "white")
p2
ggsave(filename = "Galaxy_UMP.pdf", width = 7.8, height = 6.5, plot = p2)
结果如下:

PPT修饰
这里没有找到很好的办法加上细胞标签,用ppt好了,最终结果如下:
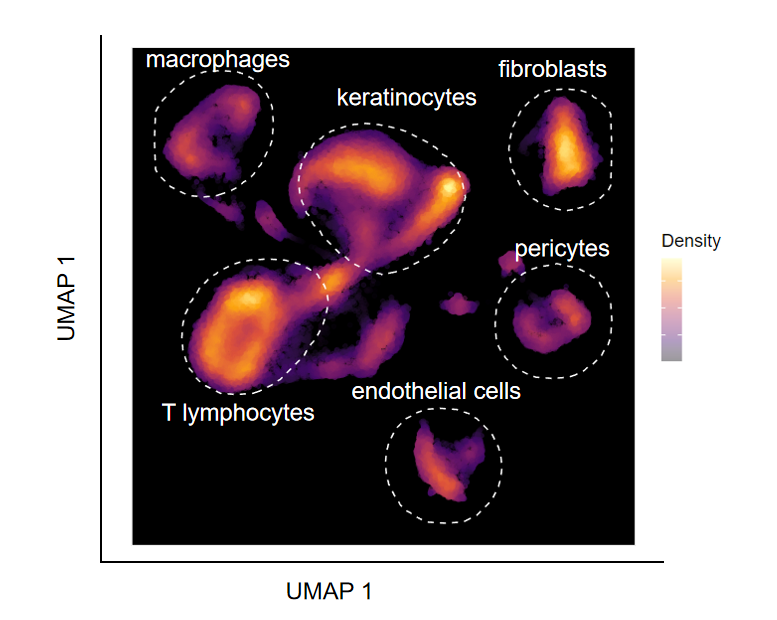
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-02-24,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

