🤩 Seurat | 空间转录组数据分析的标准流程!~(一)(数据处理与整合分析)
🤩 Seurat | 空间转录组数据分析的标准流程!~(一)(数据处理与整合分析)
生信漫卷
发布于 2025-11-13 18:24:52
发布于 2025-11-13 18:24:52
写在前面
随着空间转录组学的发展,我们不仅能看到基因在细胞里表达多少,还能够真正回答这些细胞究竟在哪里的问题。😅
📍 在组织切片上,不同细胞类型的分布就像一座城市的街区划分:有的细胞成片聚集,有的只在某个区域安家。
今天是Seurat分析空转的教程,一起学习吧!~😍
用到的包
rm(list = ls())
library(Seurat)
library(SeuratData)
library(ggplot2)
library(patchwork)
library(dplyr)
示例数据
这里,我们使用Visium v1的矢状小鼠脑切片数据集。🧐
有两个连续的anterior和两个(匹配的)连续posterior。🐶
#InstallData("stxBrain")
brain <- LoadData("stxBrain", type = "anterior1")
brain

数据预处理
我们首先需要对数据进行归一化,以考虑数据点之间测序深度的差异。😁
对于空间数据集,spot的方差可能很大,特别是当整个组织的细胞密度存在差异时。我们在这里看到了很大的异质性,这需要有效的归一化。😘
plot1 <- VlnPlot(brain, features = "nCount_Spatial", pt.size = 0.1) + NoLegend()
plot2 <- SpatialFeaturePlot(brain, features = "nCount_Spatial") + theme(legend.position = "right")
wrap_plots(plot1, plot2)
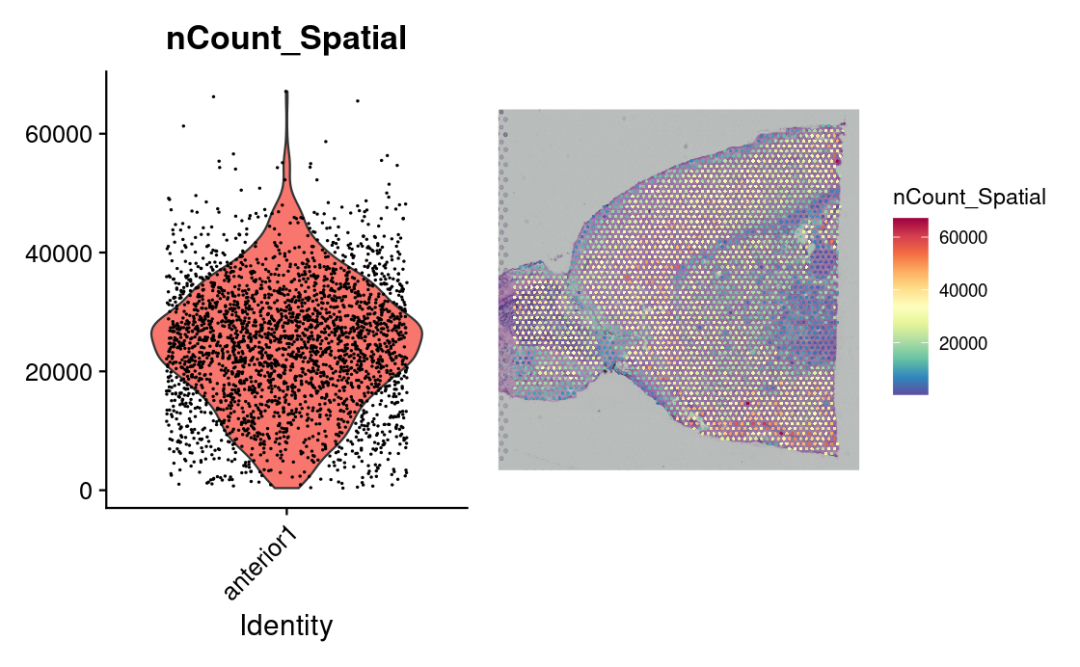
标准方法(如LogNormalize()函数),会强制每个数据点在归一化后具有相同的底层size,可能会有问题。🫠
所以这里我们推荐使用sctransform构建基因表达的正则化负二项式模型,以便在保留生物方差的同时解释技术伪影。🙊
brain <- SCTransform(brain, assay = "Spatial", verbose = FALSE)
基因表达可视化
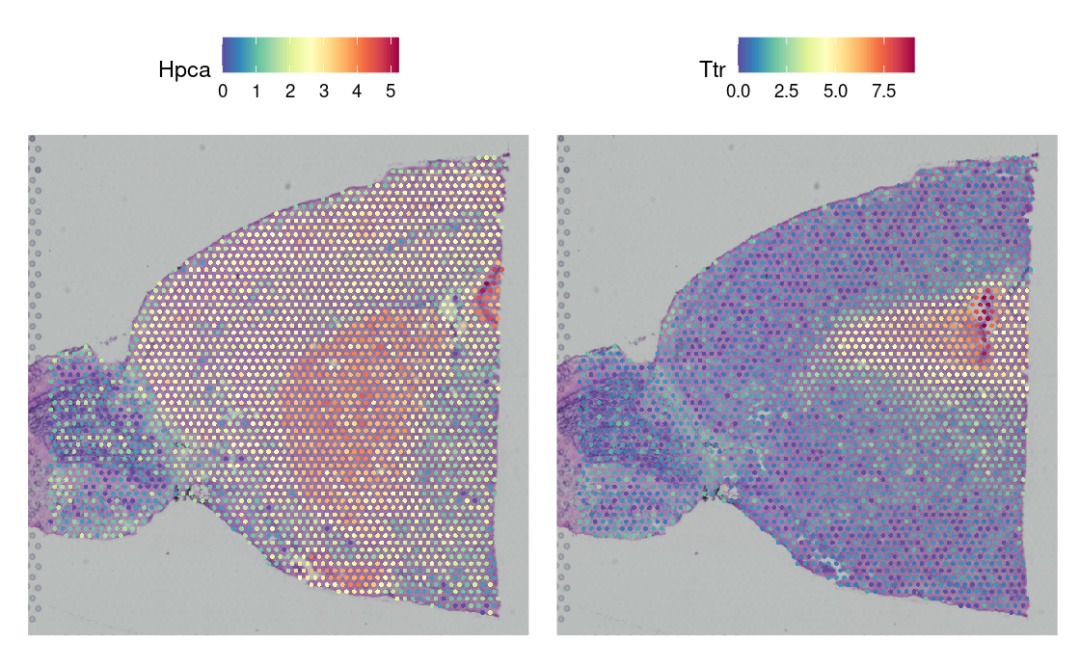
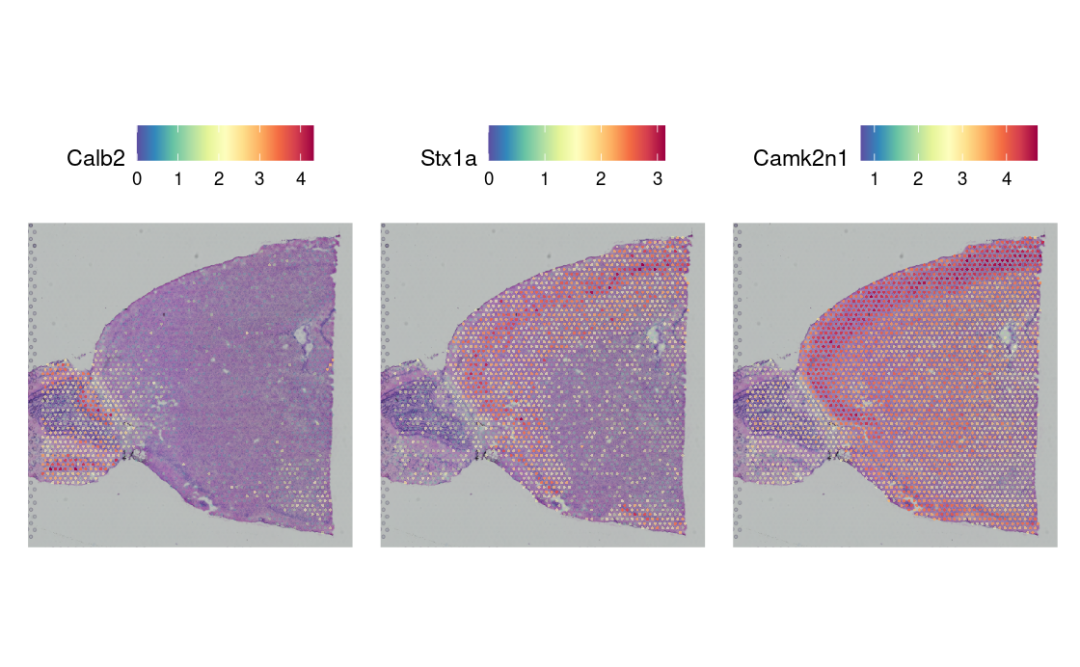
在这个小鼠大脑的数据集中,基因Hpca是强海马体marker,Ttr是脉络丛的marker。😁
SpatialFeaturePlot(brain, features = c("Hpca", "Ttr"))
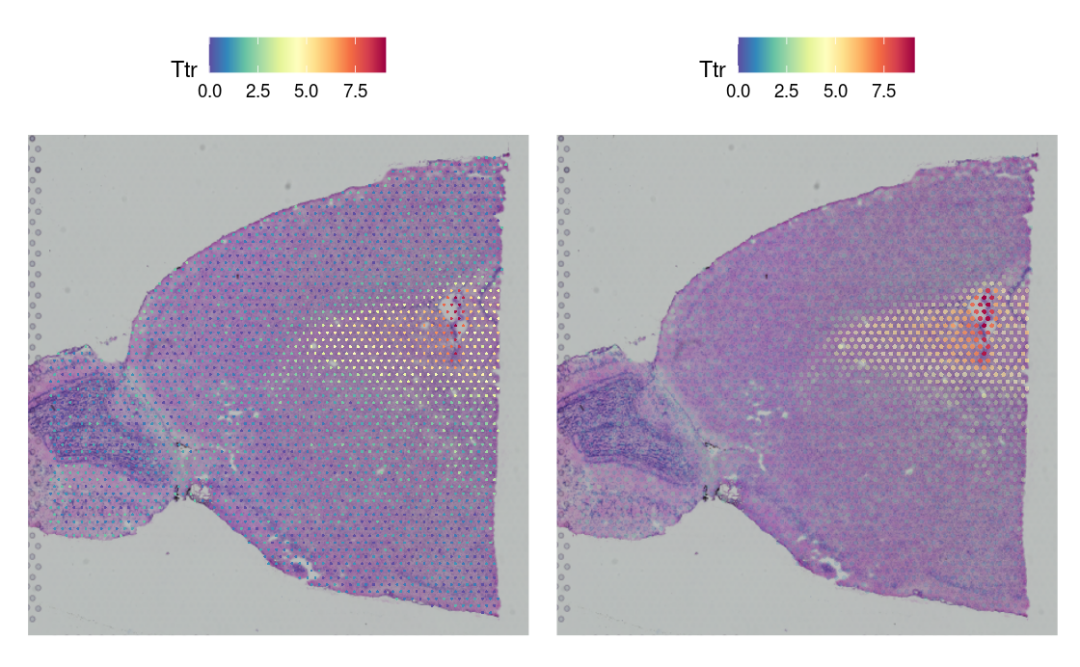
优化可视化!~
p1 <- SpatialFeaturePlot(brain, features = "Ttr", pt.size.factor = 1)
p2 <- SpatialFeaturePlot(brain, features = "Ttr", alpha = c(0.1, 1))
p1 + p2
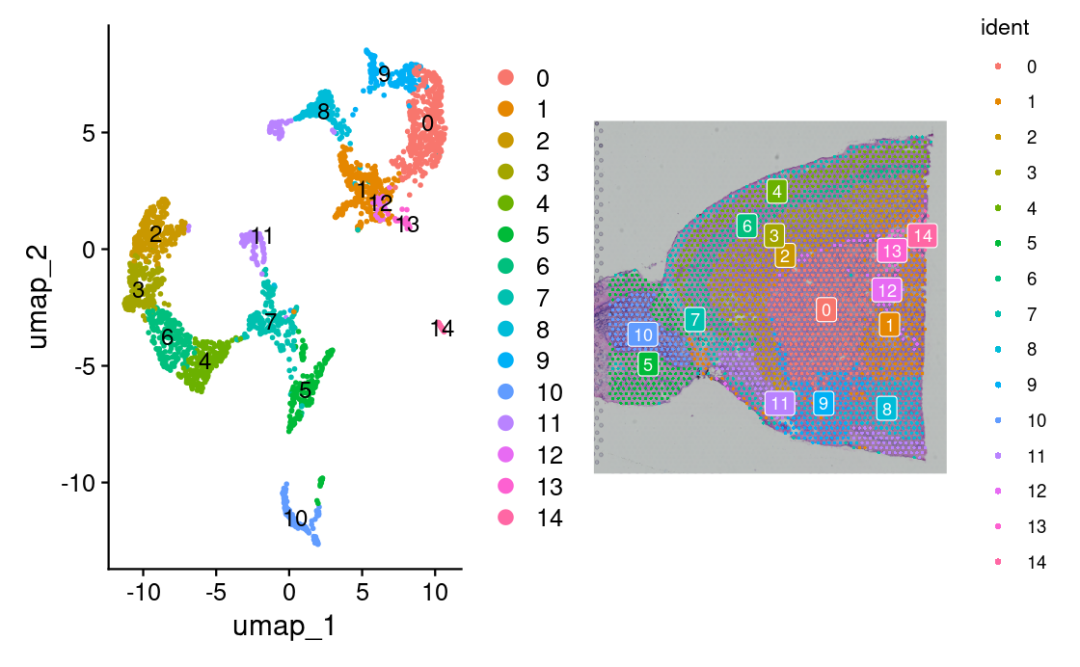
降维、聚类和可视化
我们继续使用与scRNA-seq分析相同的pipeline对RNA表达数据进行降维和聚类。😏
brain <- RunPCA(brain, assay = "SCT", verbose = FALSE)
brain <- FindNeighbors(brain, reduction = "pca", dims = 1:30)
brain <- FindClusters(brain, verbose = FALSE)
brain <- RunUMAP(brain, reduction = "pca", dims = 1:30)
p1 <- DimPlot(brain, reduction = "umap", label = TRUE)
p2 <- SpatialDimPlot(brain, label = TRUE, label.size = 3)
p1 + p2
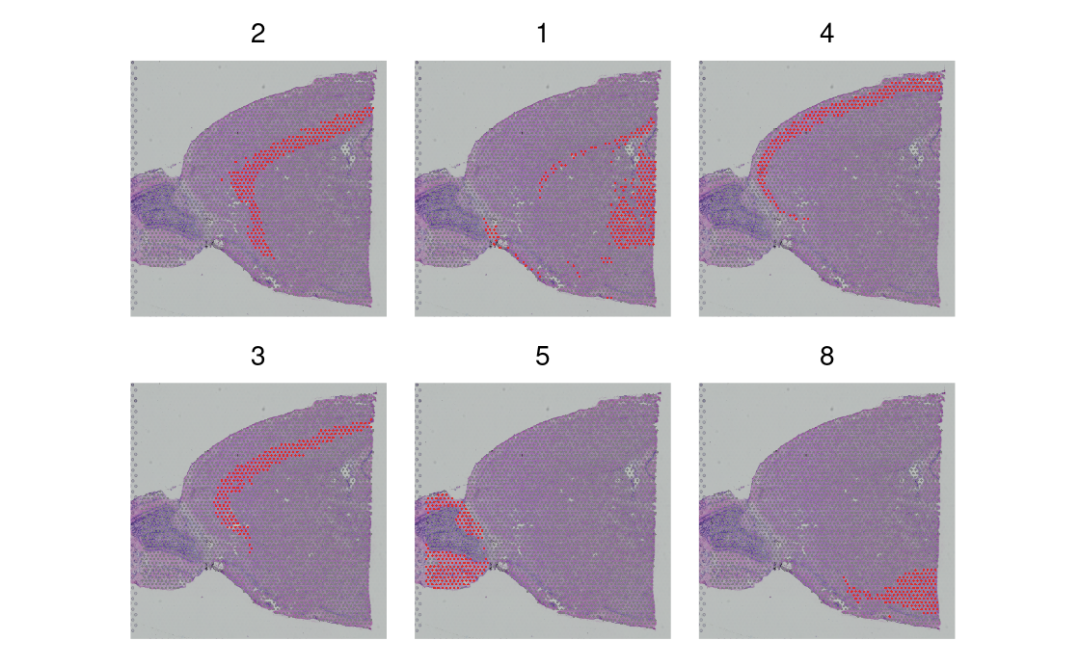
highlight某几个亚群!~⭐️
SpatialDimPlot(brain, cells.highlight = CellsByIdentities(object = brain, idents = c(2, 1, 4, 3,
5, 8)), facet.highlight = TRUE, ncol = 3)
识别空间特征基因
方法一
使用FindMarkers, 需要有一定的先验知识。🙊
de_markers <- FindMarkers(brain, ident.1 = 5, ident.2 = 6)
SpatialFeaturePlot(object = brain, features = rownames(de_markers)[1:3], alpha = c(0.1, 1), ncol = 3)
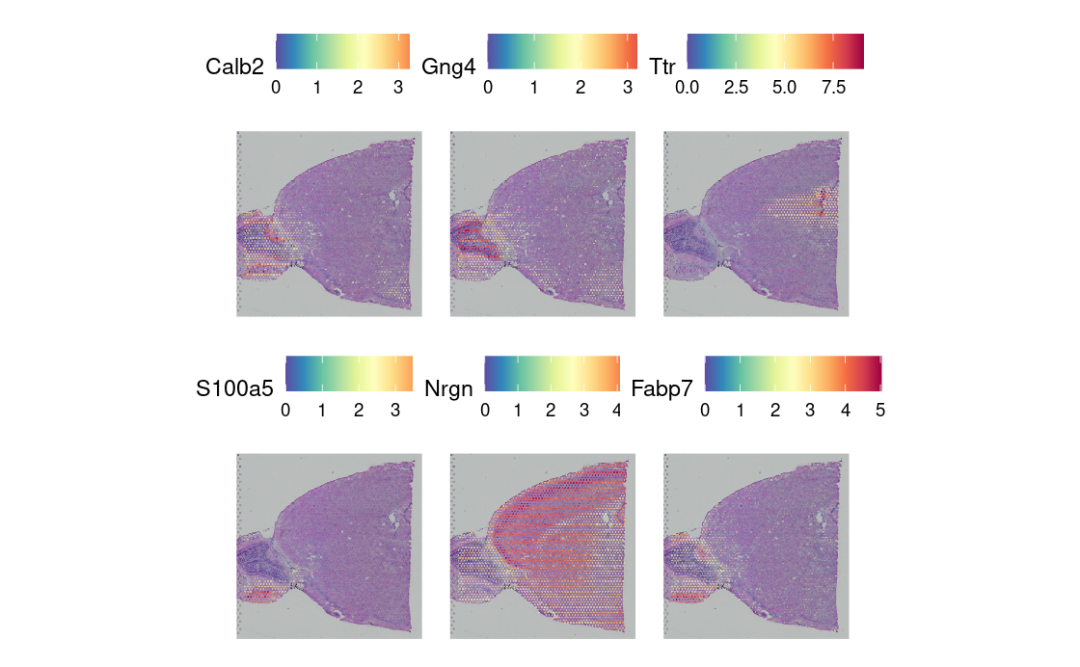
方法二
使用FindSpatiallyVariableFeatures,在没有预先注释的情况下探索空间特征。😁
这里我们可视化前6个特征基因的的表达。🥳
brain <- FindSpatiallyVariableFeatures(brain,
assay = "SCT",
features = VariableFeatures(brain)[1:1000],
selection.method = "moransi"
)
top.features <- SpatiallyVariableFeatures(brain,
method = "moransi" # “markvariogram”, “moransi”
) %>%
head()
SpatialFeaturePlot(brain, features = top.features, ncol = 3, alpha = c(0.1, 1))
感兴趣区域的深入分析
这里需要注意的是,Seurat v5后,空间坐标不再直接存储在metadata中,而是位于spatial image structure中。🫣
在进行空间过滤前,我们需要提取x和y的坐标信息,并手动添加到metadata中。🤓
# Subset to clusters of interest
cortex <- subset(brain, idents = c(1, 2, 3, 4, 6, 7))
# Extract and attach spatial coordinates from anterior1 image
centroids <- cortex[["anterior1"]]@boundaries$centroids
coords <- setNames(as.data.frame(centroids@coords), c("x", "y"))
rownames(coords) <- centroids@cells
cortex$x <- coords[colnames(cortex), "x"]
cortex$y <- coords[colnames(cortex), "y"]
# Perform spatial subsetting using image position Subset to upper right quadrant
cortex <- subset(cortex, y < 2719 | x > 7835, invert = TRUE)
# Further remove cells in lower right quadrant
m <- (9395 - 5747)/(4960 - 7715)
b <- 5747 - m * 7715
cortex <- subset(cortex, y > m * x + b, invert = TRUE)
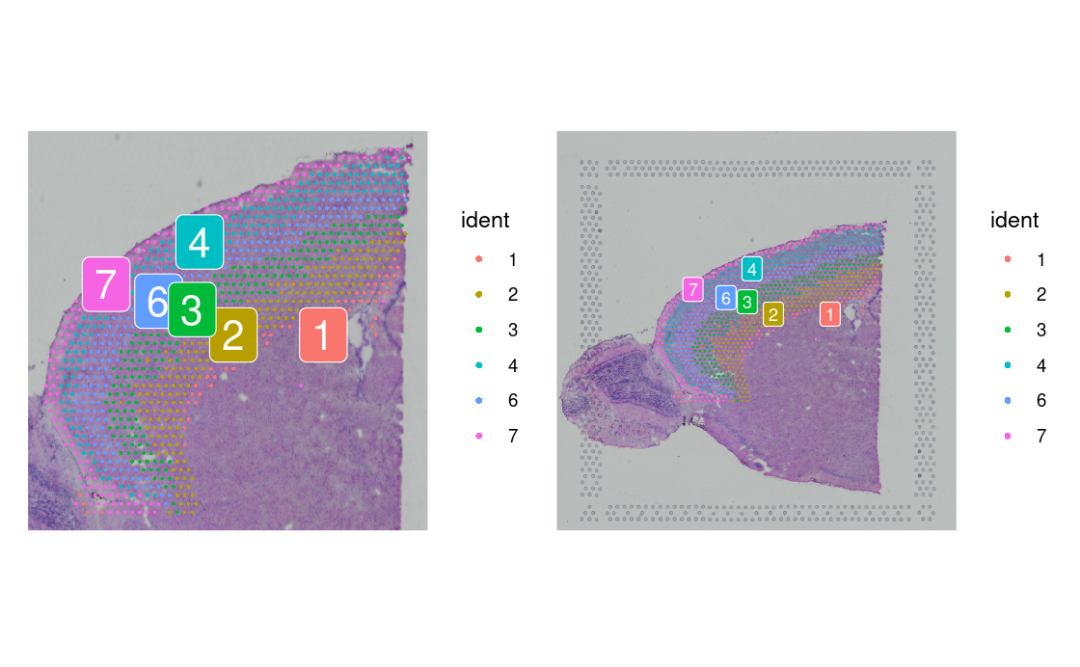
# Visualize the subsetted data on full image vs cropped image
p1 <- SpatialDimPlot(cortex, crop = TRUE, label = TRUE)
p2 <- SpatialDimPlot(cortex, crop = FALSE, label = TRUE, pt.size.factor = 1, label.size = 3)
p1 + p2
与单细胞数据整合
在~50um时,visium的spot将包含多个细胞的表达谱。🤪
我们可以使用scRNA-seq数据,反卷积每个spot以预测细胞类型的潜在组成。😘
allen_reference <- readRDS("./Seurat_spatial/allen_cortex.rds")
allen_reference
这里我们使用anchor的方法来整合单细胞数据和空间数据。🥸(官方认为这种方法比反卷积效果更好!~)
library(dplyr)
library(future)
plan(multisession, workers = 4)
options(future.globals.maxSize = 16 * 1024^3)
allen_reference <- SCTransform(allen_reference, ncells = 3000, verbose = FALSE) %>%
RunPCA(verbose = FALSE) %>%
RunUMAP(dims = 1:30)
sctransform等常规操作!~😘
# After subsetting, we renormalize cortex
cortex <- SCTransform(cortex, assay = "Spatial", verbose = FALSE) %>%
RunPCA(verbose = FALSE)
# the annotation is stored in the 'subclass' column of object metadata
DimPlot(allen_reference, group.by = "subclass", label = TRUE)
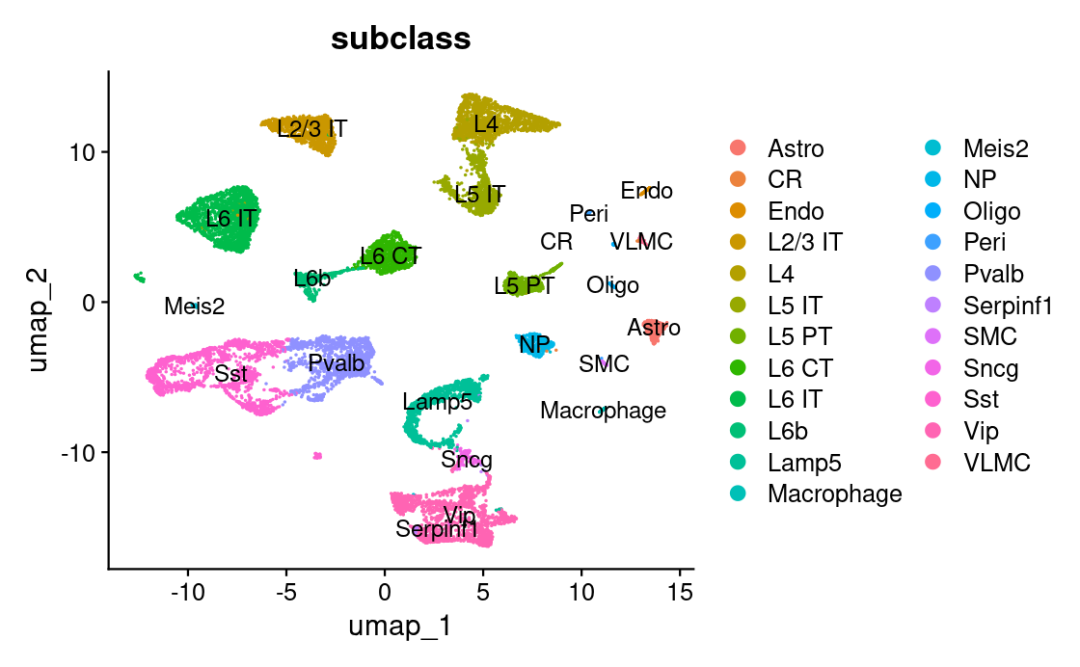
现在我们得到每种细胞在每个spot的预测分数。🥰
anchors <- FindTransferAnchors(reference = allen_reference, query = cortex, normalization.method = "SCT")
predictions.assay <- TransferData(anchorset = anchors, refdata = allen_reference$subclass, prediction.assay = TRUE,
weight.reduction = cortex[["pca"]], dims = 1:30)
cortex[["predictions"]] <- predictions.assay
在这里,我们可以区分这些神经元亚型的分布位置。😛
DefaultAssay(cortex) <- "predictions"
SpatialFeaturePlot(cortex, features = c("L2/3 IT", "L4"), pt.size.factor = 1.6, ncol = 2, crop = TRUE)
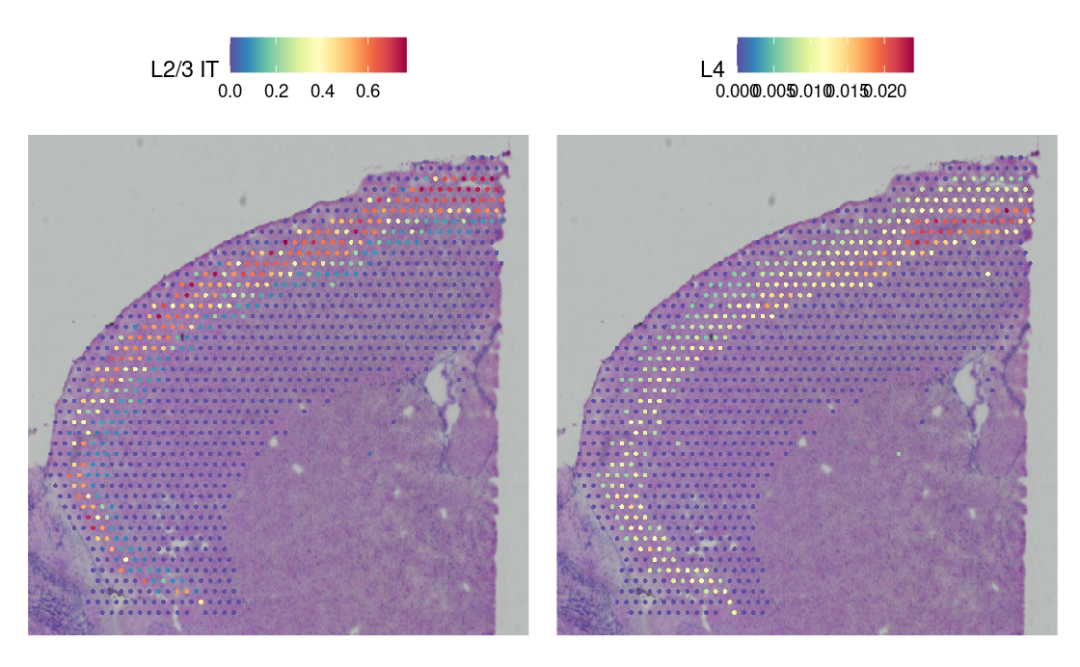
我们先算出了每个细胞属于某种类型的可能性分数。😀
然后,借助这些分数,我们还能看出哪些细胞类型只会出现在组织的某些特定区域。🤓
做法和我们平时找‘在空间上变化显著的基因’很像,只不过这次拿来当分析依据的不是基因表达量,而是‘细胞类型预测分数’。🥳
cortex <- FindSpatiallyVariableFeatures(cortex, assay = "predictions", selection.method = "moransi",
features = rownames(cortex), r.metric = 5, slot = "data")
top.clusters <- head(SpatiallyVariableFeatures(cortex, selection.method = "moransi"), 4)
SpatialPlot(object = cortex, features = top.clusters, ncol = 2)
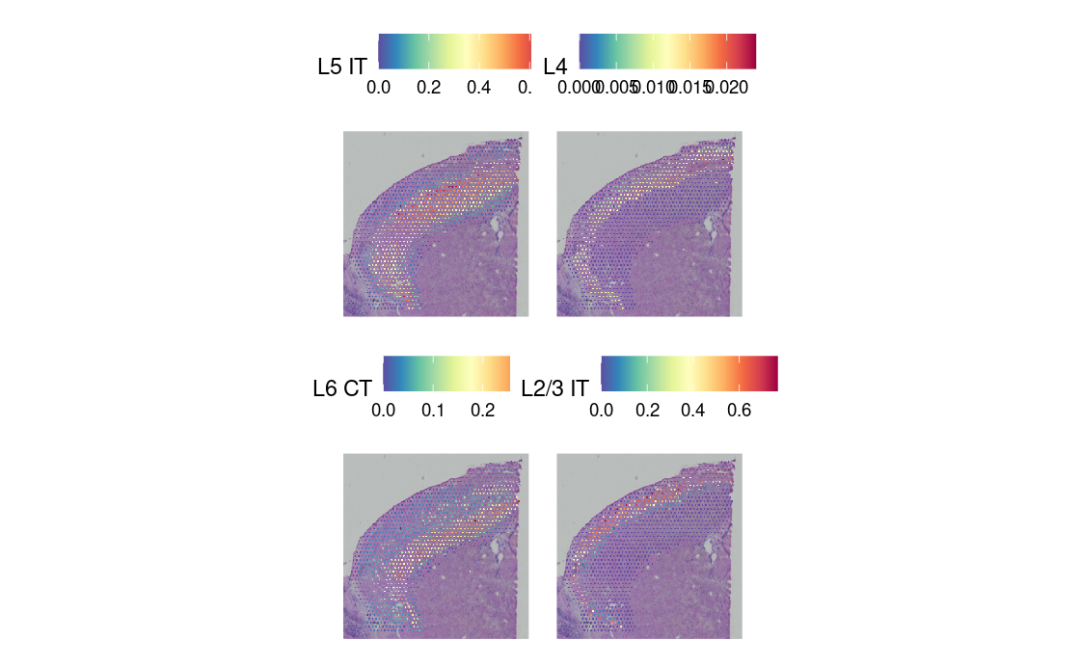
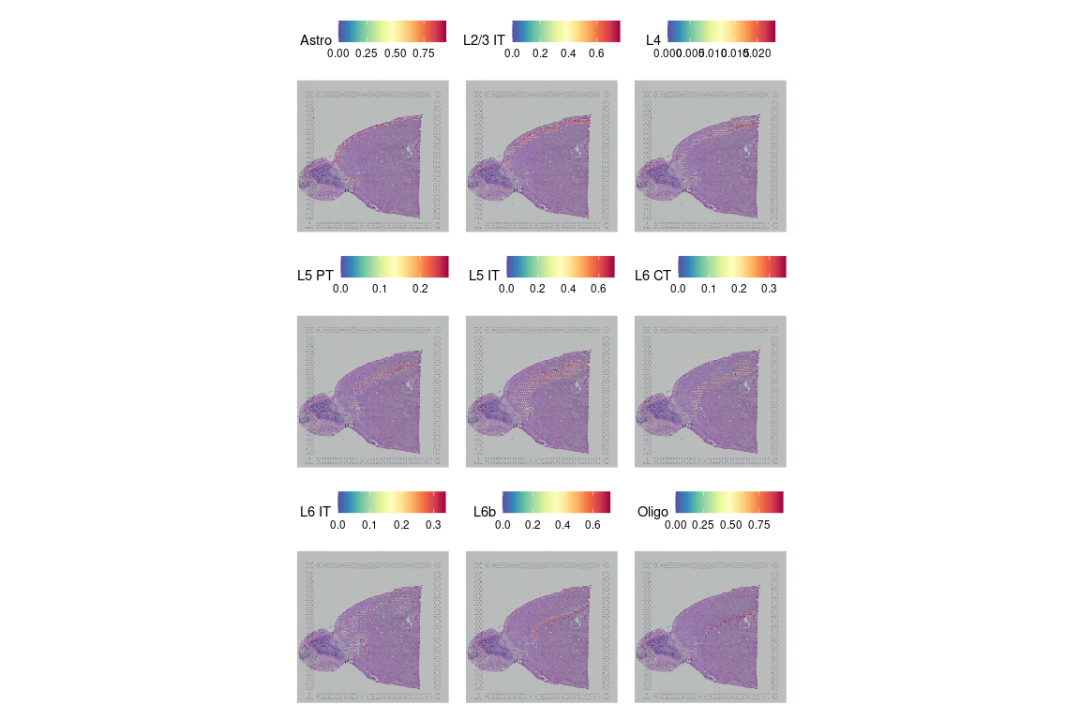
最后,我们证明这种整合分析的方法确实管用:它能把大家早就知道的那些空间分布规律‘找回来’。😛
比如,不同层里的兴奋性神经元、分布在第一层的星形胶质细胞,还有大脑皮层的灰质区域,这些都能被准确识别出来。🫢
SpatialFeaturePlot(cortex, features = c("Astro", "L2/3 IT", "L4", "L5 PT", "L5 IT", "L6 CT", "L6 IT",
"L6b", "Oligo"), pt.size.factor = 1, ncol = 3, crop = FALSE, alpha = c(0.1, 1))
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-09-07,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

