使用python读取疑难杂症的单细胞数据(GSM7870858)
使用python读取疑难杂症的单细胞数据(GSM7870858)
生信技能树
发布于 2025-11-20 11:31:04
发布于 2025-11-20 11:31:04
数据情况
先来看看数据:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM7870858
来自数据集 GSE246536,对4例健康对照患者的胎盘、4例妊娠晚期有症状COVID-19患者的胎盘,以及1例妊娠中期感染有症状COVID-19但在康复后足月采集的胎盘进行了sci-RNA-seq3分析。
提供的文件如下:
GSM7870858_105.23_cds.RDS.gz 11.5 Mb (ftp)(http) RDS
GSM7870858_105.23_cell_annotations.txt.gz 692.6 Kb (ftp)(http) TXT
GSM7870858_105.23_gene_annotations.txt.gz 291.0 Kb (ftp)(http) TXT
GSM7870858_105.23_umi_counts.mtx.gz 18.6 Mb (ftp)(http) MTX
有个rds的,R的数据读起来很方便,这里就不多说了。关键是我们现在需要使用python处理数据。
看看三个gz文件里面的内容
文件:cell_annotations.txt.gz
这个是单纯的一列barcode 细胞id:
zless GSM7870858_105.23_cell_annotations.txt.gz | wc -l # 290331
zless -S GSM7870858_105.23_cell_annotations.txt.gz |head
#
# E02_A01_P02-E04_LIG1
# E02_A01_P02-E04_LIG100
# E02_A01_P02-E04_LIG101
# E02_A01_P02-E04_LIG103
# E02_A01_P02-E04_LIG105
# E02_A01_P02-E04_LIG108
# E02_A01_P02-E04_LIG109
# E02_A01_P02-E04_LIG112
# E02_A01_P02-E04_LIG114
# E02_A01_P02-E04_LIG115
文件:gene_annotations.txt.gz
基因信息:两列,tab分割
zelss GSM7870858_105.23_gene_annotations.txt.gz |wc -l # 43945
less -S GSM7870858_105.23_gene_annotations.txt.gz |head
# ENSG00000000003 TSPAN6
# ENSG00000000005 TNMD
# ENSG00000000419 DPM1
# ENSG00000000457 SCYL3
# ENSG00000000460 C1orf112
# ENSG00000000938 FGR
# ENSG00000000971 CFH
# ENSG00000001036 FUCA2
# ENSG00000001084 GCLC
# ENSG00000001167 NFYA
文件:umi_counts.mtx.gz
标准的单细胞数据矩阵
zless -S GSM7870858_105.23_umi_counts.mtx.gz
# %%MatrixMarket matrix coordinate integer general
# %
# 43945 290331 5864791
# 42237 1 1
# 1558 2 1
# 21175 3 1
# 32766 3 1
# 28384 4 1
# 5322 5 1
# 7562 6 1
# 3553 7 1
# 4806 7 1
# 632 8 1
# 2214 9 1
上面三个文件就是cellranger定量完了之后的标准文件,但是名字还需要改一下。标准的三个文件:
- barcodes.tsv.gz
- features.tsv.gz
- matrix.mtx.gz
mkdir matrix
cp GSM7870858_105.23_cell_annotations.txt.gz matrix/barcodes.tsv.gz
cp GSM7870858_105.23_gene_annotations.txt.gz matrix/features.tsv.gz
cp GSM7870858_105.23_umi_counts.mtx.gz matrix/matrix.mtx.gz
scanpy读取
改完名字后:

读取就非常简单啦:先导入模块
import pandas as pd
import scanpy as sc
import numpy as np
sc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)
sc.logging.print_header() #查看主要包库的版本
sc.settings.set_figure_params(dpi=80, facecolor="white")
scanpy的函数:
adata = sc.read_10x_mtx(
"matrix/", # the directory with the `.mtx` file
var_names="gene_symbols", # use gene symbols for the variable names (variables-axis index)
cache=True, # write a cache file for faster subsequent reading
)
但是报错啦:(果然没有这么容易!)
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File /nas2/zhangj/biosoft/miniconda3/envs/sc/lib/python3.10/site-packages/pandas/core/indexes/base.py:3812, in Index.get_loc(self, key)
3811 try:
-> 3812 return self._engine.get_loc(casted_key)
3813 except KeyError as err:
File pandas/_libs/index.pyx:167, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/index.pyx:196, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/hashtable_class_helper.pxi:2606, in pandas._libs.hashtable.Int64HashTable.get_item()
File pandas/_libs/hashtable_class_helper.pxi:2630, in pandas._libs.hashtable.Int64HashTable.get_item()
KeyError: 2
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
Cell In[3], line 1
----> 1 adata = sc.read_10x_mtx(
2 "matrix/", # the directory with the `.mtx` file
3 var_names="gene_symbols", # use gene symbols for the variable names (variables-axis index)
4 cache=True, # write a cache file for faster subsequent reading
5 )
File /nas2/zhangj/biosoft/miniconda3/envs/sc/lib/python3.10/site-packages/legacy_api_wrap/__init__.py:82, in legacy_api.<locals>.wrapper.<locals>.fn_compatible(*args_all, **kw)
79 @wraps(fn)
80 def fn_compatible(*args_all: P.args, **kw: P.kwargs) -> R:
81 if len(args_all) <= n_positional:
---> 82 return fn(*args_all, **kw)
84 args_pos: P.args
85 args_pos, args_rest = args_all[:n_positional], args_all[n_positional:]
File /nas2/zhangj/biosoft/miniconda3/envs/sc/lib/python3.10/site-packages/scanpy/readwrite.py:572, in read_10x_mtx(path, var_names, make_unique, cache, cache_compression, gex_only, prefix)
570 prefix = ""if prefix is None else prefix
571 is_legacy = (path / f"{prefix}genes.tsv").is_file()
--> 572 adata = _read_10x_mtx(
573 path,
574 var_names=var_names,
575 make_unique=make_unique,
576 cache=cache,
577 cache_compression=cache_compression,
578 prefix=prefix,
579 is_legacy=is_legacy,
580 )
581 if is_legacy or not gex_only:
582 return adata
File /nas2/zhangj/biosoft/miniconda3/envs/sc/lib/python3.10/site-packages/scanpy/readwrite.py:622, in _read_10x_mtx(path, var_names, make_unique, cache, cache_compression, prefix, is_legacy)
620 raise ValueError(msg)
621 if not is_legacy:
--> 622 adata.var["feature_types"] = genes[2].values
623 barcodes = pd.read_csv(path / f"{prefix}barcodes.tsv{suffix}", header=None)
624 adata.obs_names = barcodes[0].values
File /nas2/zhangj/biosoft/miniconda3/envs/sc/lib/python3.10/site-packages/pandas/core/frame.py:4107, in DataFrame.__getitem__(self, key)
4105 if self.columns.nlevels > 1:
4106 return self._getitem_multilevel(key)
-> 4107 indexer = self.columns.get_loc(key)
4108 if is_integer(indexer):
4109 indexer = [indexer]
File /nas2/zhangj/biosoft/miniconda3/envs/sc/lib/python3.10/site-packages/pandas/core/indexes/base.py:3819, in Index.get_loc(self, key)
3814 if isinstance(casted_key, slice) or (
3815 isinstance(casted_key, abc.Iterable)
3816 and any(isinstance(x, slice) for x in casted_key)
3817 ):
3818 raise InvalidIndexError(key)
-> 3819 raise KeyError(key) from err
3820 except TypeError:
3821 # If we have a listlike key, _check_indexing_error will raise
3822 # InvalidIndexError. Otherwise we fall through and re-raise
3823 # the TypeError.
3824 self._check_indexing_error(key)
KeyError: 2
上面这个报错看起来有点多,我直接扔给人工智能deepseek解读:
这个错误是因为您的 10x Genomics 数据文件格式与 Scanpy 期望的格式不匹配。错误发生在尝试读取
genes.tsv文件时,Scanpy期望该文件至少有3列(基因ID、基因符号、特征类型),但您的文件可能只有2列。
确实,我检查了一下 标准的genes.tsv 文件有三列:
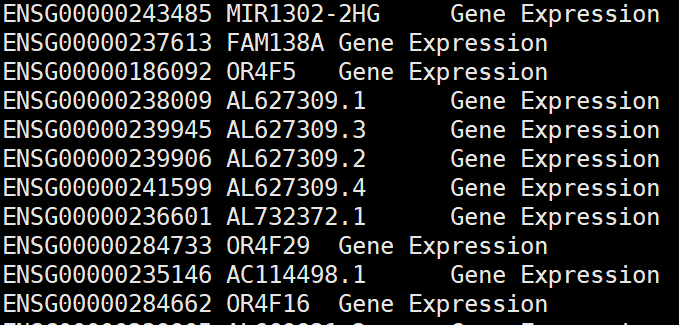
改一下:在原来的文件后面新增一列 Gene Expression
cp features.tsv.gz features.tsv.gz.bak
zless features.tsv.gz.bak |sed 's/$/\tGene Expression/' >features.tsv
gzip features.tsv
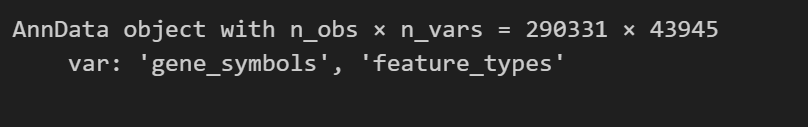
这下可以了:
adata = sc.read_10x_mtx( "matrix/", var_names="gene_symbols", cache=True)
adata.var_names_make_unique()
# this is unnecessary if using `var_names='gene_ids'` in `sc.read_10x_mtx`
adata
现在学习生信,遇到报错也不用害怕了,你相当于有了一个24小时陪练在线的高级工程师给你进行各种指导!
当然,如果还有人工智能也解决不了的,来找生信技能树吧!
下面,我构建了一个针对生信初学者学习的微信群,会发布《生信人如何系统入门R 2025版》,《生信人如何系统入门python 2025版》、《生信人如何系统入门Linux 2025版》的相关学习资料带领大家学习,帮助你跨过那个陌生的第一步!
本文参与 腾讯云自媒体同步曝光计划,分享自微信公众号。
原始发表:2025-11-19,如有侵权请联系 cloudcommunity@tencent.com 删除
评论
登录后参与评论
推荐阅读
目录

