【从零学习OpenCV 4】极坐标变换
【从零学习OpenCV 4】极坐标变换
经过几个月的努力,小白终于完成了市面上第一本OpenCV 4入门书籍《从零学习OpenCV 4》。为了更让小伙伴更早的了解最新版的OpenCV 4,小白与出版社沟通,提前在公众号上连载部分内容,请持续关注小白。
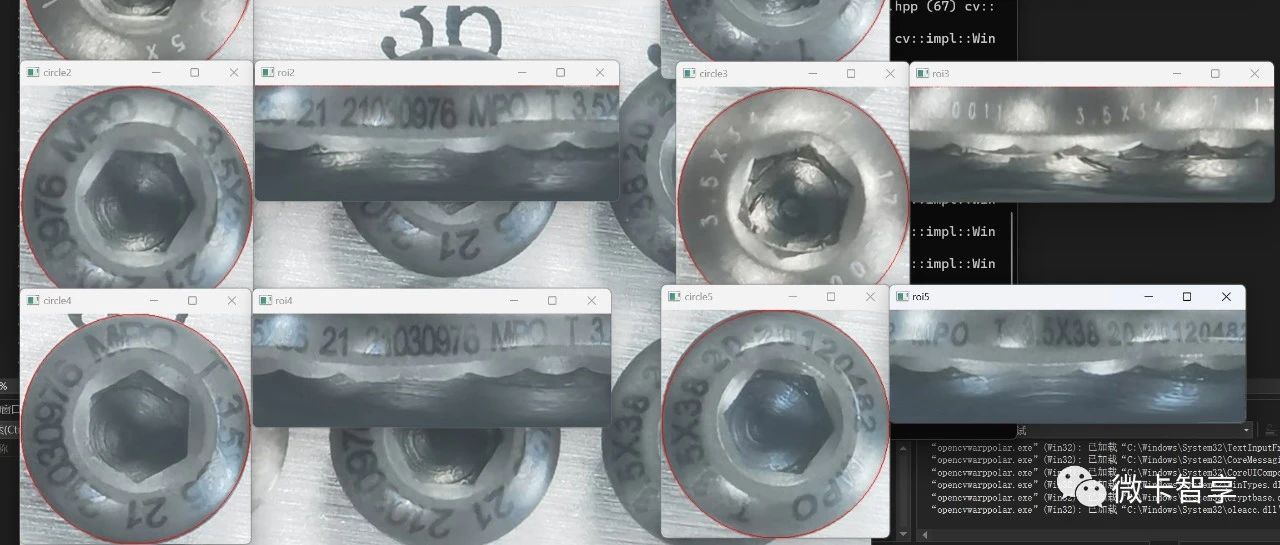
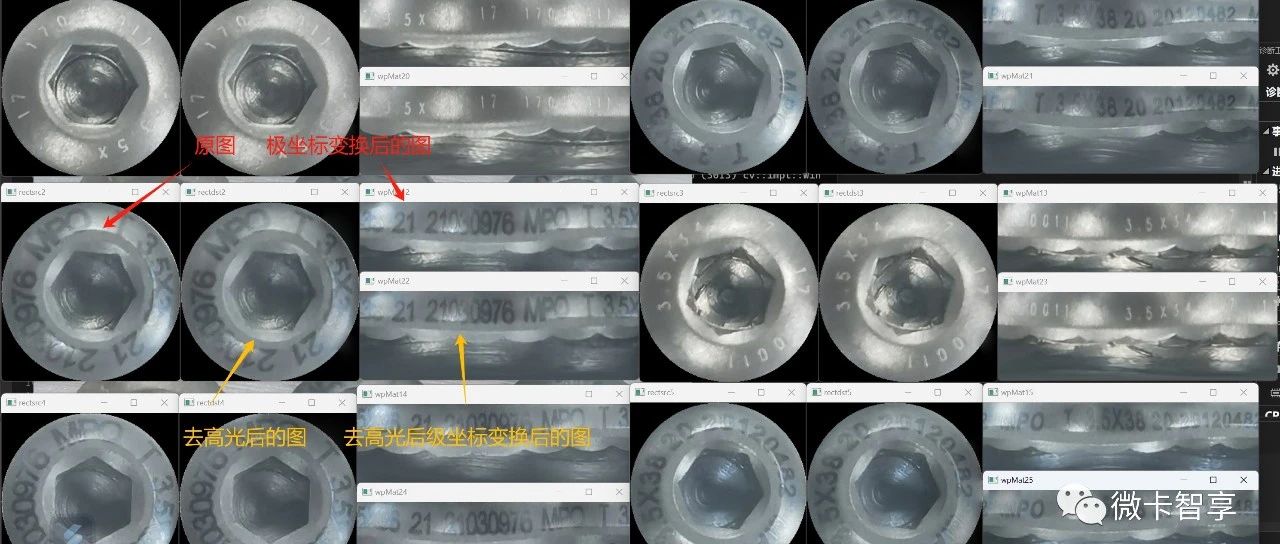
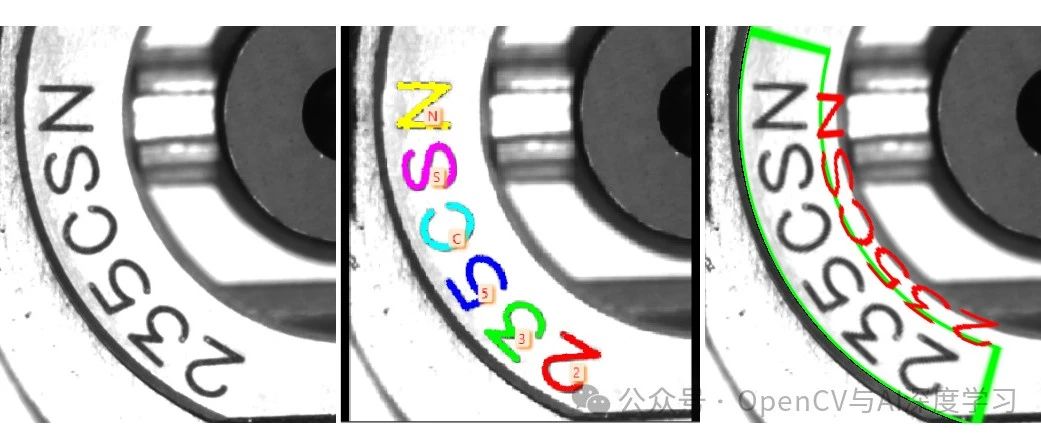
极坐标变换就是将图像在直角坐标系与极坐标系中互相变换,形式如图3-26所示,它可以将一圆形图像变换成一个矩形图像,常用于处理钟表、圆盘等图像。圆形图案边缘上的文字经过及坐标变换后可以垂直的排列在新图像的边缘,便于对文字的识别和检测。

图3-26 极坐标变换示意图
OpenCV 4中提供了warpPolar()函数用于实现图像的极坐标变换,该函数的函数原型在代码清单3-38中给出。
代码清单3-38 warpPolar()函数原型
1. void cv::warpPolar(InputArray src,
2. OutputArray dst,
3. Size dsize,
4. Point2f center,
5. double maxRadius,
6. int flags
7. )- src:原图像,可以是灰度图像或者彩色图像。
- dst:极坐标变换后输出图像,与原图像具有相同的数据类型和通道数。
- dsize:目标图像大小。
- center:极坐标变换时极坐标的原点坐标。
- maxRadius:变换时边界圆的半径,它也决定了逆变换时的比例参数。
- flags: 插值方法与极坐标映射方法标志,插值方法在表3-3中给出,极坐标映射方法在表3-7给出,两个方法之间通过“+”或者“|”号进行连接。
该函数实现了图像极坐标变换和半对数极坐标变换。函数第一个参数是需要进行极坐标变换的原始图像,该图像可以是灰度图像也可以是彩色图像。第二个参数是变换后的输出图像,与输入图像具有相同的数据类型和通道数。第三个参数是变换后图像的大小。第四个参数是极坐标变换时极坐标原点在原图像中的位置,该参数同样适用于逆变换中。第五个参数是变换时边界圆的半径,它也决定了逆变换时的比例参数。最后一个参数是变换方法的选择标志,插值方法在表3-3中给出,极坐标映射方法在表3-7给出,两个方法之间通过“+”或者“|”号进行连接。
表3-7 warpPolar()函数极坐标映射方法标志
标志参数 | 作用 |
|---|---|
WARP_POLAR_LINEAR | 极坐标变换 |
WARP_POLAR_LOG | 半对数极坐标变换 |
WARP_INVERSE_MAP | 逆变换 |
该函数可以对图像进行极坐标正变换也可以进行逆变换,关键在于最后一个参数如何选择。为了了解图像极坐标变换的功能以及相关函数的使用,在代码清单3-39给出了对表盘图像进行极坐标正变换和逆变换的示例程序。程序中选取表盘的中心作为极坐标的原点,变换的结果在图3-27给出。
代码清单3-39 mywarpPolar.cpp图像极坐标变换
1. #include <opencv2\opencv.hpp>
2. #include <iostream>
3.
4. using namespace std;
5. using namespace cv;
6.
7. int main()
8. {
9. Mat img = imread("dial.png");
10. if (!img.data)
11. {
12. cout << "请检查图像文件名称是否正确" << endl;
13. return -1;
14. }
15.
16. Mat img1, img2;
17. Point2f center = Point2f(img.cols / 2, img.rows/2); //极坐标在图像中的原点
18. //正极坐标变换
19. warpPolar(img, img1, Size(300,600), center, center.x,
20. INTER_LINEAR + WARP_POLAR_LINEAR);
21. //逆极坐标变换
22. warpPolar(img1, img2, Size(img.rows,img.cols), center, center.x,
23. INTER_LINEAR + WARP_POLAR_LINEAR + WARP_INVERSE_MAP);
24.
25. imshow("原表盘图", img);
26. imshow("表盘极坐标变换结果", img1);
27. imshow("逆变换结果", img2);
28. waitKey(0);
29. return 0;
30. }
图3-27 mywarpPolar.cpp程序中极坐标正变换和逆变换的结果
腾讯云开发者

扫码关注腾讯云开发者
领取腾讯云代金券
Copyright © 2013 - 2025 Tencent Cloud. All Rights Reserved. 腾讯云 版权所有
深圳市腾讯计算机系统有限公司 ICP备案/许可证号:粤B2-20090059 深公网安备号 44030502008569
腾讯云计算(北京)有限责任公司 京ICP证150476号 | 京ICP备11018762号 | 京公网安备号11010802020287
Copyright © 2013 - 2025 Tencent Cloud.
All Rights Reserved. 腾讯云 版权所有